Review: KRAS mutations are influential in driving hepatic metastases and predicting outcome in colorectal cancer
Introduction
Colorectal cancer (CRC) remains a significant healthcare concern and remains the third most common cause of cancer-related death in the USA and is the 2nd leading cause of death (1). Death from CRC is usually due to metastatic disease, and how to best treat, stratify, and improve treatments for metastatic CRC (mCRC) patients using biomarkers to guide precise treatment decisions is a topic of intense study and interest. A better understanding of the biology of CRC has led to some advances in oncological therapies for patients. Cetuximab is an epidermal growth factor inhibitor which plays a role in treatment of mCRC. There have been numerous randomised controlled trials assessing the efficacy of the addition of anti-EGFR to current first-line chemotherapy with varying results. The greatest benefits have been seen in patients with Kirsten Rat sarcoma viral oncogene (KRAS) wild type tumors. KRAS mutations have been shown to exert a negative effect on efficacy of cetuximab when used with other chemotherapy agents (2).
Despite these advances, surgery remains the most effective therapy in the setting of oligometastatic disease and in particular, colorectal liver metastases (3). This review focuses on our understanding of the important role that KRAS mutations play in the progression of colorectal cancer. We describe the frequencies of KRAS mutations, their relevance to CRC in the metastatic setting, and proposed mechanisms by which these mutations may drive metastasis. We examine how future trials may look to target KRAS mutant tumors.
KRAS mutation in colorectal cancer
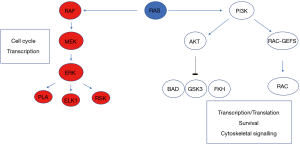
RAS proteins control signalling pathways that in the presence of normal homeostatic mechanisms lead to controlled cell growth and proliferation (4). Knockout mouse studies have shown that Kras deletion leads to synthetic lethality and, unlike closely related proteins Hras and Nras, is required for normal development (5). In the setting of cancer activating oncogenic mutations, usually via point mutations, there is chronic ‘switching-on’ of these pathways and resultant malignant potential (6). Activated RAS protein expression leads to dysregulated tumour cell growth, invasiveness and angiogenesis (7). KRAS mutations activate signal transduction via GTP binding. By binding RAF, activated KRAS sets in motion a sequence of kinase activation that results in MEK then sequentially ERK kinase activation, resulting in ERK translocation to the nucleus and engagement of transcription mechanisms (Figure 1). Additionally, PI3-kinase activation may be engaged, resulting in control of a large number of downstream enzymes. It was recognised as early as 1987 by Bert Vogelstein’s group that KRAS was mutated in approximately 30% of cases and mutations appeared most commonly at codon 12 (8). CRC was comprehensively molecularly characterised in 2012 (9). KRAS was mutated in 43% of non-hypermutated cases of CRC, with mutations of the oncogenic codons 12 and 13 of exon 2 predominating. Indeed, almost all RAS activity, in tumours from different tissues of origin, is due to activating mutations in codons 12, 13, and 61 (10). Data obtained from trials suggests chemotherapy resistance mechanisms may also be mediated by mutations in exon 4, codon 146, though the influence of codon 61 and 146 mutations is difficult to ascertain from studies due to their relative rarity. Interestingly, 55% of non-hypermutated patients had alterations in KRAS, NRAS, or BRAF with the majority of these being mutually exclusive. Furthermore, Poulin et al. (11) have recently shown by utilising a cross-disciplinary approach that tissue-specific phenotypes (i.e., those specific to the colon) result from the signalling properties of each KRAS allele. This work suggests that engagement of signalling mechanisms downstream of KRAS mutations is both tissue and context dependent.
Concordance in KRAS mutations between primary and metastatic sites in colorectal cancer
As described, mutant KRAS functions as a transducer of growth signals downstream of EGFR activation following constitutive activation of G12D/G13 codons, resulting in the essential neoplastic function of KRAS mutations, and subsequent resistance of tumor cells to EGFR inhibitors including cetuximab (12). Therefore, KRAS status of tumors has played an important role in determining subsequent sensitivity to EGFR inhibitors. Likewise, microsatellite instability (MSI) and mismatch repair protein expression segregates a subset of patients that may respond favourably to T cell directed checkpoint inhibition including PD1 inhibitors (13,14). Tissue is required for assessment of mutation status. Studies have assessed correlation between primary and metastatic sites, analysing if primary tissue is sufficient to guide treatment. Fujiyoshi et al. enrolled 457 patients with primary and metastatic tumours (499 synchronous and 57 metachronous metastases) and performed analysis of KRAS and BRAF mutation and MSI status (15). Concordance was very high between primary and metastatic site and remained high across all mutations (>90%). KRAS status between primary and metastatic site showed discordance in 16 cases. Most of these cases were discordant between lymph node metastases and may be due to intratumoral heterogeneity. Liver metastases showed high concordance, in keeping with Knijn et al. (16). These data confirm KRAS mutations are an early event in colorectal carcinogenesis. There was no difference observed between synchronous and metachronous metastasis formation.
Bhullar et al. published a meta-analysis of biomarker concordance between primary and metastatic sites in CRC (17). They confirmed across all studies KRAS concordance was 93.7%. These findings are in keeping with Vakiani et al. who found very high concordance between primary and secondary sites for KRAS mutations but not TP53 and BRAF mutations (18). These data confirm that sampling of either primary or metastatic site is adequate for assessment of KRAS mutations that are key in treatment decision making and in helping understand the process of metastasis in CRC. Few genetic changes are thought to occur in progression from primary to metastatic disease, suggesting that epigenetic and microenvironmental factors are likely to play a key role in the process of metastasis in CRC.
Clinical relevance: KRAS influence on prognosis following liver resection
Liver resection remains the only option for cure for patients with CRLM (19,20). Extent of disease involvement is the primary measure of concern when making treatment decisions to ensure limitation of postoperative morbidity and maximisation of survival benefit (21). Prediction of early recurrence remains difficult. Modern treatment regimens now dictate that preoperative chemotherapy is standard of care prior to CRLM resection. Medical oncologists use KRAS status to select patients for EGFR-directed chemotherapy in more advanced, irresectable disease. However, KRAS mutation status represents a useful prognostic marker in response to therapy for patients undergoing liver resection and can be incorporated into modern risk scores to help inform treatment decision making. Brudvik et al. undertook a meta-analysis of studies examining the influence of KRAS mutations on outcome following CRLM resection. In the 14 studies that met inclusion criteria, 8 studies associated KRAS status with overall survival (22). KRAS mutations were adversely associated with overall survival following liver resection in each of these studies, and with recurrence free survival in 7 of these. These findings are entirely independent of EGFR inhibitor utilisation. These data demonstrate in a clinical context that the relevance of KRAS mutations in CRC extends beyond response to EGFR inhibitors and has an important role to play in progression of advanced disease. Indeed, this study identified the propensity for higher rates of KRAS mutation in widespread metastatic disease, with patients with resectable oligometastatic disease harbouring fewer KRAS mutations than most studies of all-comers with metastatic disease. These findings must be considered in light of findings by certain authors who have found traditional clinical risk scores limited in their usefulness when assessing response to resection for CRLM (23). The main advantage of KRAS mutation status in this context is as a single biomarker, an early event in colorectal cancer pathogenesis, and an event unperturbed by chemotherapy. Importantly though, recent work by Kawaguchi et al. has demonstrated that KRAS status alone carries less prognostic power than KRAS mutations coexisting with mutations in TP53 and SMAD4 (24). In future, personalised therapies will look to target complex interactions between multiple genetic aberrations in colorectal cancer.
The relationship between KRAS expression, mutation location and outcome following resection of colorectal liver metastases
It is accepted that KRAS and BRAF mutations are associated with poor prognosis following resection for CRLM. Interestingly, using a temporal assessment formula, Margonis et al. have shown that as time passes from resection, surgeon dependent factors including margin status and resected extrahepatic disease predominate after the first year (25). By using a meta-analysis approach, Dr. Pawlik and his group have shown that KRAS is a poor prognostic factor following CRLM resection, with BRAF mutant patients having even poorer prognosis (26). Additionally, in response to preoperative chemotherapy and low CRLM tumor burden, 5-year survival for KRAS/NRAS/BRAF WT patients was 55%, KRAS/NRAS mutant patients was 35%, and for BRAF mutant patients was 0%. Therefore, genetic status, tumor burden and subsequent change in tumor burden should influence pre- and post-operative chemotherapy decision making in the setting of CRLM resection (27).
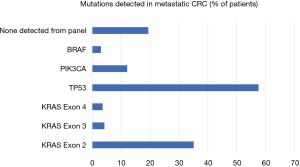
Given that resistance to chemotherapy regimens may be mediated by genetic heterogeneity within tumors, investigators have begun to assess the impact of mutation location on outcome following resection for CRLM. Frankel et al. studied the location of KRAS mutation in context of survival following resection for CRLM (28). Whole genome analysis of 211 patients revealed that 43% had either a KRAS or NRAS mutation with the majority found in KRAS. Most mutations were in codons 12 and 13 with the majority in G12D, S or V and G13C or D. Exon 3 (codon 61) accounted for 10% of mutations and exon 4 (codon 146) 8.5% (Figure 2). There were clear differences between exon 3 and 4 tumors. Exon 4 tumors were larger, more likely to be solitary, and associated with longer disease-free survival. Kwak et al. performed meta-analysis of codon 13 mutations on overall survival; they confirmed both codons 12 and 13 were associated with poorer overall survival compared with wild type tumors and there were no differences between the 2 groups (29). Interestingly, when patients were separated based on EGFR therapies, those that received EGFR therapy had similar outcomes if they harboured codon 13 mutations or were WT. Such meta-analyses do have limitations, as other mutation status and confounding factors are not controlled for in these trials (e.g., number of mutations samples, NGS platform used). Recently Margonis et al. have confirmed that KRAS mutations were associated with poorer response to pre-hepatectomy chemotherapy; however, codon 13 mutations correlated with the worst responses, poorer than codon 12. These data suggest differences in biology between sites of mutation and subsequent differences in resistance to chemotherapy (30).
Interestingly, in the Frankel study, pattern of recurrence was also influenced by genotype. BRAF mutant patients suffered multifocal recurrence and KRAS mutant patients recurred in liver and lung in a similar fashion to wild type patients. Vauthey et al. similarly captured all KRAS mutations and found KRAS mutation was associated with poorer outcome following resection of CRLM (31). Of note, their data were striking for the low level of KRAS mutations within their cohort but this could simply be due to selection or referral bias. Interestingly, in a separate cohort of CRLM patients that had hepatic artery infusion chemotherapy, KRAS status did not influence outcome, suggesting this technique may overcome some of the aggressive features of KRAS mutant disease (32). The literature however is seeded with studies that have analysed only exon 2 mutations, some of these have shown poorer overall survival in KRAS mutant patients (33,34), while others (35) have observed no influence. Importantly, the patients in Frankel’s cohort demonstrate that up to 20% of KRAS mutations are harboured in exons 3 and 4 and therefore must be considered when assessing KRAS status, as these mutations convey very different recurrence and survival profiles. Data from Janakiraman et al. confirm this finding that exon 4 mutant tumors are larger, have less malignant potential when compared with exon 2 and 3 mutations (36) and correlate with functional studies in mice performed by Poulin et al. (11) that show different phenotypes generated as a result of G12D (exon 2) or A146T (exon 4) mutations. Future studies should adopt a broader next generation sequencing platform to permit detection of all KRAS mutations and to consider them in the context of all genetic aberrations in individual patients to permit personalisation of therapies.
Increasing complexity: KRAS association with stage and molecular subtypes of disease
Following early sequencing genomic sequencing studies, investigators have begun to focus less on individual mutations and on characterisation of the pattern of mutations and gene expression to subclassify CRC. Guinney et al. subclassified CRC into the consensus molecular subtypes (CMS) in 2015 in an effort to resolve inconsistencies in assessment of the disease and to promote clinical targeting and translation (37). KRAS mutant tumours were distributed throughout the 4 subtypes but were overrepresented in CMS 3 disease.
CALGB/SWOG 80405 assessed patients with metastatic colorectal cancer treated with first line therapy plus either bevacizumab or cetuximab. The study showed no significant difference in overall survival between groups. Innocenti et al. in their analysis of mutations in the trial showed that patients with tumours with high mutational burden survived longer, while patients with both BRAF and KRAS mutant tumours had poor overall survival (38). Williams et al. (39) looked at early colorectal tumours and again found that those with high mutational burden survived longest; similarly to findings by Van den Eynde and colleagues (40), they showed that tumour infiltrating lymphocyte response was the greatest predictor of outcome. Immune infiltration superseded dMMR, pMMR/BRAFwt/KRASwt, pMMR/BRAFmut/KRASwt, pMMR/BRAFwt/KRASmut profiles and indeed transcriptional CMS subtypes in predicting outcome. Lal et al. (41) showed that KRAS mutation and CMS2 and 3 were independently associated with reduced immune cell infiltration, associated with poorer outcome. Pitroda et al. (42) found that liver metastases when considered in isolation by CMS RNA signatures >30% were unable to be classified accurately. They derived their own signature that showed once again that KRAS was strongly associated with the worst subtype of disease. Furthermore, they derived an immune subtype associated with the most favourable outcomes and a canonical subtype that predicted intermediate outcomes. These studies suggest that mutant KRAS is associated with the most aggressive disease and provides the stimulus for development of an aggressive, stromal/mesenchymal microenvironment that promotes metastatic progression as described in recent murine modelling studies.
At present and as evidenced by the MRC COIN trial, there is a large gap in our understanding as to the mechanism of resistance that underlies EGFR-directed therapies, and optimum combination therapies for patients with advanced CRC (43). This large randomised controlled trial showed no difference in progression free and overall survival in patients receiving first line palliative oxaliplatin plus or minus cetuximab in both KRAS wild type and mutant patients. Indeed, a recent phase II trial (44) has demonstrated that patients carrying polymorphisms in non-functional receptor KIR2DS4 respond positively to cetuximab therapy. The only group that seemed to benefit in COIN were those wild type patients on infusion fluorouracil and cetuximab, with limited disease. One positive factor from COIN was the stratification in response seen between KRAS mutant and wild type patients, with BRAF mutant patients faring worst of all. Future trials will look to stratify patient subsets based on molecular subtypes, while it is clear that novel strategies and targets within the microenvironment of metastases need to be trialled to improve understanding of the disease and resistance mechanisms. One hundred eighty-eight trials have recently recruited, or are ongoing, using KRAS as a biomarker in metastatic colorectal cancer. These trials, though too numerous to list, give an appreciation of the importance of KRAS to the future understanding of metastatic CRC and in developing appropriate therapies for patients (clinicaltrials.gov).
Mechanism of KRAS-driven metastasis
Recent murine modelling studies have provided an insight into the role of mutant KRAS in generating and maintaining CRC metastases. Using an inducible Kras allele (exon 2, G12D) in combination with conditional null Apc and Tp53 alleles, the ‘iKAP’ model, Boutin et al. showed that the model faithfully progressed from adenoma to metastasis when Kras was mutated, but not in the presence of wild type Kras (45). Exome sequencing revealed that mutant Kras cell populations were homogenous in metastases while heterogeneous in primary tumours suggesting the dependency of metastases on this subpopulation of cells. Genetic elimination of mutant Kras led to absence of this population at primary and secondary sites and subsequent apoptotic elimination of cells, suggesting that mutant Kras is important for maintenance of CRC metastases. High levels of TGF-β expression were seen, in keeping with a mesenchymal phenotype and consistent with strong overlap with the most invasive CMS4 CRC seen in humans. Incredibly when mutant Kras was switched off in advanced tumors, these tumors reverted to a benign adenomatous morphology.
De Pinho’s group have also recently demonstrated that Kras mutant cells engage myeloid derived suppressor cells via CXCR2 signalling which then promote immunosuppression and evasion of checkpoint blockade (46). This mechanism is believed to be reliant on interferon regulatory factor 2. Similarly, our group have observed an important role of CXCR2-positive MDSCs in driving metastases in mutant Kras CRC. Our findings suggest Notch signalling, downstream of tumour CXCL5 expression, plays a critical role in engagement of these cells and subsequent immunosuppression (Jackstadt et al., In Press, Cancer Cell). The importance of these observations lies in resistance to immunotherapy seen in CRC (47). Elimination of T cell suppressive cells from these tumours in the future may render them amenable to targeting by immunotherapy (48).
Additionally, Chu et al. have demonstrated that activation of IGF-1R downstream of mutant Kras leads to reversible induction of metastasis in murine models, raising the possibility of a role for MEK inhibition in a subset of patients with metastatic CRC (49). Batlle’s group has shown on an Apc, Kras mutant background that TGF-β produced by tumor stroma regulates immunogenicity by influencing T cell responses. In experimental models TGF-β inhibition released this block on T cell responses and sensitised metastases to PD1 checkpoint inhibition (50), indicating a potential therapeutic strategy for future trials. In this setting it will be interesting to observe the development of parallel tumor biopsies and serial ctDNA monitoring to assess response to therapy and assessment of reasons for resistance to specifically targeted therapies (51). Indeed, Dr. Corcoran and his group have already shown that tumor heterogeneity can drive molecular resistance to therapy and influence lesion-specific responses to targeted therapies (52).
Conclusions
KRAS mutant CRC patients have poorer disease free and overall survival. Patients tend to develop aggressive tumours that metastasize with high concordance in mutation status between primary and secondary sites. Insights from modelling studies suggest mutant KRAS works at multiple levels to sustain an aggressive phenotype of colorectal cancer. Recent work in human cohorts suggests, however, that the site of mutations may influence tumor behaviour and outcome. KRAS remains an important biomarker of aggressive disease, and trials have shown it can stratify response to therapy and recurrence after resection. Work into KRAS mutations in the context of the transcriptional profile of metastatic CRC is ongoing. It is likely these aggressive tumors will require multiple individually targeted therapies, in combination with real-time assessment of resistance mechanisms, to prevent recurrence in adequately resected CRLMs in the future.
Acknowledgments
None.
Footnote
Conflicts of Interest: Dr. Smith has received travel support from Intuitive Surgical Inc. for fellow education and has served as a clinical advisor for Guardant Health, Inc. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Surgeons ACo. Colorectal Cancer Facts and Figures 2017-2019. Cancer.org 2019.
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [Crossref] [PubMed]
- Barbon C, Margonis GA, Andreatos N, et al. Colorectal Liver Metastases: Does the Future of Precision Medicine Lie in Genetic Testing? J Gastrointest Surg 2018;22:1286-96. [Crossref] [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Johnson L, Greenbaum D, Cichowski K, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev 1997;11:2468-81. [Crossref] [PubMed]
- Guo F, Gong H, Zhao H, et al. Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients. Sci Rep 2018;8:6076. [Crossref] [PubMed]
- Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 2002;418:934. [Crossref] [PubMed]
- Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987;327:293-7. [Crossref] [PubMed]
- Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res 1989;49:4682-9. [PubMed]
- Poulin EJ, Bera AK, Lu J, et al. Tissue-Specific Oncogenic Activity of KRAS(A146T). Cancer Discov 2019;9:738-55. [Crossref] [PubMed]
- Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 2012;486:532-6. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Diaz LA Jr, Le DT. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;373:1979. [Crossref] [PubMed]
- Fujiyoshi K, Yamamoto G, Takahashi A, et al. High concordance rate of KRAS/BRAF mutations and MSI-H between primary colorectal cancer and corresponding metastases. Oncol Rep 2017;37:785-92. [Crossref] [PubMed]
- Knijn N, Mekenkamp LJ, Klomp M, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer 2011;104:1020-6. [Crossref] [PubMed]
- Bhullar DS, Barriuso J, Mullamitha S, et al. Biomarker concordance between primary colorectal cancer and its metastases. EBioMedicine 2019;40:363-74. [Crossref] [PubMed]
- Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956-62. [Crossref] [PubMed]
- Andreou A, Aloia TA, Brouquet A, et al. Margin status remains an important determinant of survival after surgical resection of colorectal liver metastases in the era of modern chemotherapy. Ann Surg 2013;257:1079-88. [Crossref] [PubMed]
- House MG, Kemeny NE, Gonen M, et al. Comparison of adjuvant systemic chemotherapy with or without hepatic arterial infusional chemotherapy after hepatic resection for metastatic colorectal cancer. Ann Surg 2011;254:851-6. [Crossref] [PubMed]
- Margonis GA, Sasaki K, Gholami S, et al. Genetic And Morphological Evaluation (GAME) score for patients with colorectal liver metastases. Br J Surg 2018;105:1210-20. [Crossref] [PubMed]
- Brudvik KW, Kopetz SE, Li L, et al. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg 2015;102:1175-83. [Crossref] [PubMed]
- Kumar R, Dennison AR, Robertson V, et al. Clinical risk scores in the current era of neoadjuvant chemotherapy for colorectal liver metastases. ANZ J Surg 2018;88:E16-20. [Crossref] [PubMed]
- Kawaguchi Y, Kopetz S, Newhook TE, et al. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Margonis GA, Buettner S, Andreatos N, et al. Prognostic Factors Change Over Time After Hepatectomy for Colorectal Liver Metastases: A Multi-institutional, International Analysis of 1099 Patients. Ann Surg 2019;269:1129-37. [Crossref] [PubMed]
- Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol 2018;27:280-8. [Crossref] [PubMed]
- Ruzzenente A, Bagante F, Ratti F, et al. Response to preoperative chemotherapy: impact of change in total burden score and mutational tumor status on prognosis of patients undergoing resection for colorectal liver metastases. HPB (Oxford) 2019. [Epub ahead of print].
- Frankel TL, Vakiani E, Nathan H, et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer 2017;123:568-75. [Crossref] [PubMed]
- Kwak MS, Cha JM, Yoon JY, et al. Prognostic value of KRAS codon 13 gene mutation for overall survival in colorectal cancer: Direct and indirect comparison meta-analysis. Medicine (Baltimore) 2017;96:e7882. [Crossref] [PubMed]
- Margonis GA, Amini N, Andreatos N, et al. KRAS mutational status impacts pathologic response to pre-hepatectomy chemotherapy: a study from the International Genetic Consortium for Liver Metastases. HPB (Oxford) 2019. [Epub ahead of print].
- Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013;258:619-26; discussion 26-7. [Crossref] [PubMed]
- Gholami S, Kemeny NE, Boucher TM, et al. Adjuvant Hepatic Artery Infusion Chemotherapy is Associated With Improved Survival Regardless of KRAS Mutation Status in Patients With Resected Colorectal Liver Metastases: A Retrospective Analysis of 674 Patients. Ann Surg 2019. [Epub ahead of print].
- Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137-44. [Crossref] [PubMed]
- Kemeny NE, Chou JF, Capanu M, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer 2014;120:3965-71. [Crossref] [PubMed]
- Etienne-Grimaldi MC, Mahamat A, Chazal M, et al. Molecular patterns in deficient mismatch repair colorectal tumours: results from a French prospective multicentric biological and genetic study. Br J Cancer 2014;110:2728-37. [Crossref] [PubMed]
- Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res 2010;70:5901-11. [Crossref] [PubMed]
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med 2015;21:1350-6. [Crossref] [PubMed]
- Innocenti F, Ou FS, Qu X, et al. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol 2019;37:1217-27. [Crossref] [PubMed]
- Williams DS, Mouradov D, Jorissen RN, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut 2018. [Epub ahead of print]. [PubMed]
- Van den Eynde M, Mlecnik B, Bindea G, et al. The Link between the Multiverse of Immune Microenvironments in Metastases and the Survival of Colorectal Cancer Patients. Cancer Cell 2018;34:1012-26.e3. [Crossref] [PubMed]
- Lal N, White BS, Goussous G, et al. KRAS Mutation and Consensus Molecular Subtypes 2 and 3 Are Independently Associated with Reduced Immune Infiltration and Reactivity in Colorectal Cancer. Clin Cancer Res 2018;24:224-33. [Crossref] [PubMed]
- Pitroda SP, Khodarev NN, Huang L, et al. Integrated molecular subtyping defines a curable oligometastatic state in colorectal liver metastasis. Nat Commun 2018;9:1793. [Crossref] [PubMed]
- Adams RA, Meade AM, Seymour MT, et al. Intermittent versus continuous oxaliplatin and fluoropyrimidine combination chemotherapy for first-line treatment of advanced colorectal cancer: results of the randomised phase 3 MRC COIN trial. Lancet Oncol 2011;12:642-53. [Crossref] [PubMed]
- Borrero-Palacios A, Cebrian A, Gomez Del Pulgar MT, et al. Combination of KIR2DS4 and FcgammaRIIa polymorphisms predicts the response to cetuximab in KRAS mutant metastatic colorectal cancer. Sci Rep 2019;9:2589. [Crossref] [PubMed]
- Boutin AT, Liao WT, Wang M, et al. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 2017;31:370-82. [Crossref] [PubMed]
- Liao W, Overman MJ, Boutin AT, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019;35:559-72.e7. [Crossref] [PubMed]
- Basile D, Garattini SK, Bonotto M, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin Biol Ther 2017;17:709-21. [Crossref] [PubMed]
- Morton JP, Sansom OJ. CXCR2 inhibition in pancreatic cancer: opportunities for immunotherapy? Immunotherapy 2017;9:9-12. [Crossref] [PubMed]
- Chu PC, Lin PC, Wu HY, et al. Mutant KRAS promotes liver metastasis of colorectal cancer, in part, by upregulating the MEK-Sp1-DNMT1-miR-137-YB-1-IGF-IR signaling pathway. Oncogene 2018;37:3440-55. [Crossref] [PubMed]
- Tauriello DVF, Palomo-Ponce S, Stork D, et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018;554:538-43. [Crossref] [PubMed]
- Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov 2016;6:147-53. [Crossref] [PubMed]
- Oddo D, Sennott EM, Barault L, et al. Molecular Landscape of Acquired Resistance to Targeted Therapy Combinations in BRAF-Mutant Colorectal Cancer. Cancer Res 2016;76:4504-15. [Crossref] [PubMed]