
Clinical features and mutation analysis of class 1/2/3 BRAF mutation colorectal cancer
Highlight box
Key findings
• Patients with BRAF (B-Raf proto-oncogene, serine/threonine kinase) mutation had significant association with high tumor mutational burden (TMB-H) and high microsatellite instability (MSI-H), especially in class 1 BRAF mutation.
• Class 1 are more likely to co-occur with passenger mutation.
• Class 1 was mutually exclusive with co-KRAS mutation and co-APC mutation appeared more frequently in non-class 1.
• Median overall survival of class 1/2/3/not available (NA) (unknown) was 19.43/28.50/47.57 months/not reached (P=0.0001), respectively.
What is known and what is new?
• BRAF inhibitor (BRAFi) or BRAFi + EGFR inhibitor/MEK inhibitor have notably improved survival in BRAF-mutated solid tumors, however, the sensitivity in colorectal cancer (CRC) is to lower. No article reported BRAF-mutated concomitant mutation in CRC.
• We analyze class 1/2/3/NA BRAF mutation prevalence base on classification system, investigate co-mutation and pathway enrichment in Chinese and Western cohort, and compared the mutation characteristics in two cohort.
What is the implication, and what should change now?
• Class 1 may benefit from BRAFi + immunotherapy due to TMB-H and MSI. Understanding BRAF mutation types and co-mutation mechanism will promote the development of precision therapy for CRC in the future.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause for cancer-related deaths worldwide (1,2). In China, CRC ranks fifth among the most frequent malignancies in both incidence and mortality, accounting for more than two-thirds of all cancer cases (3). The efficacy of targeted therapies in CRC patients is largely unpredictable due to intrinsic genetic complexity, immune microenvironment, and partially unknown reason, although significant progress has been made in the development of mutation-driven targeted therapies in CRC patients (4). Therefore, it is urgent to understand the deep molecular mechanisms for CRC patients and improve our understanding of cancer co-mutation feature and overall survival (OS).
BRAF (B-Raf proto-oncogene, serine/threonine kinase) as one of the major oncogenic drivers, occurring in 8–12% CRC patients worldwide and was recently reported to be as high as 20.9% in CRC patients at Beijing Hospital (5,6). More than 70% BRAF mutation occur in the kinase domain, including the most commonly observed V600E mutation in CRC (7). A study has elucidated significantly mutually exclusive between BRAF and KRAS (Kirsten rat sarcoma viral oncogene homologue) mutations (8). BRAF inhibitor (BRAFi) monotherapy or in combination with an EGFR (epidermal growth factor receptor)/MEK (mitogen-activated protein kinase kinase 1) inhibitor (EGFRi/MEKi) have notably improved survival in BRAF-mutated solid tumor (9-11). However, BRAF-mutated metastatic colorectal cancer (mCRC) has shown a marked lack of sensitivity to BRAF or MEKi monotherapy in early clinical trials (12). Combined therapy of BRAFi and MEKi/EGFRi can lead to improved clinical outcomes from SWOG S1406 and BEACON CRC study (13,14). A key mechanism is inhibition of BRAF leading to increased signaling through the EGFR based on negative feedback regulation of signaling pathways in CRC. Therefore, the combination of BRAFi and EGFRi is more beneficial (15,16). Potential prognostic markers in this field have been discovered in recent years. High BRAF allele fraction (AF, ≥2%) showed worse progression-free survival (PFS)/OS than low-BRAF AF patients (<2%), suggesting that AF is an independent prognostic factor (17). RNF43 (ring finger protein 43) mutation could predict response to anti-BRAF/EGFR combinatory therapies in BRAF V600E mCRC (18). Whole transcriptome sequencing (WTS) suggests that a subset of patients with specific molecular features may derive greater clinical benefit from triplet than doublet therapy (19). This biomarker can help tailor patients’ treatments. Whereas BRAF-mutated melanoma has better efficiency to monotherapy (11). Most of these studies focused on the 600th codon mutation of BRAF, and studies have shown that there are significant differences in PFS and OS in patients with different tumor types receiving BRAFi therapy. There are also more studies on the combination of BRAFi and immunotherapy or chemotherapy. Which one is the best? BRAF non-V600E mutation were less discussed and to the best of our knowledge, there are currently no articles reporting BRAF-mutated concomitant mutation analysis.
In 2017, a deeper classification system of BRAF mutation derived from pre-clinical models functional studies based on kinase activity, RAS dependency, dimer dependency and sensitivity to vemurafenib. According to classification system, RAS-independent kinase-activating V600 monomers are categorized as class 1, including p.V600E/K/D/R/M; RAS-independent kinase-activating dimers that are resistant to vemurafenib are categorized as class 2, including p. K601E/N/T, p. L597Q/V, p. G469A/V/R, p.G464V/E and fusions; and RAS-dependent kinase-inactivating heterodimers are categorized as class 3, which consists mainly of p. G466V/E/A, p. D594N/G/A/H and p. G596D/R (20,21). A more in-depth analysis of clinicopathological, prognostic, and co-mutational features based on the BRAF mutation classification system is still needed.
In the present study, we retrospectively analyzed the high-throughput sequencing (HTS) data of 339 Chinese CRC from Beijing Hospital to survey the prevalence of class 1/2/3/not available (NA) (unknown) BRAF mutations base on classification system, investigated mutation incidence and pathway enrichment in class 1/2/3 BRAF-mutated patients, and compared the mutation characteristics of Chinese and Western populations. We aim to find out, through this study, the differences between Chinese and Western BRAF-mutated CRC patients, and compare the co-mutation with different BRAF mutation types, to make an optimal precision regimen. We present this article in accordance with the STROBE reporting checklist (available at https://cco.amegroups.com/article/view/10.21037/cco-23-117/rc).
Methods
Patients and data collection
Archival samples obtained from 339 CRC patients who underwent targeted capture next-generation sequencing (NGS) between September 2016 to June 2022 at Beijing Hospital (Beijing, China) were included in this case-control study. Tissue samples and peripheral blood samples were collected for genomic sequencing tests and as germline controls, respectively. The clinicopathological features were collected from patient medical records, which are summarized in Table 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Hospital (No. 2023BJYYEC-103-01) and informed consent was taken from all the patients.
Table 1
| Characteristics | Total (n=339) | BRAF mutation (n=35) | BRAF wild-type (n=304) | P value |
|---|---|---|---|---|
| Age at diagnosis (years) | NA | |||
| Median | 63 | 62 | 63 | |
| Range | 25–93 | 28–87 | 25–93 | |
| Gender, n (%) | 0.6261 | |||
| Male | 189 (55.75) | 19 (54.29) | 170 (55.92) | |
| Female | 143 (42.18) | 16 (45.71) | 127 (41.78) | |
| NA | 7 (2.06) | 0 | 7 (2.30) | |
| Sample type, n (%) | 0.4463 | |||
| Primary | 230 (67.85) | 26 (74.29) | 204 (67.11) | |
| Metastasis | 66 (19.47) | 4 (11.43) | 62 (20.39) | |
| NA | 43 (12.68) | 5 (14.29) | 38 (12.50) | |
| Primary tumor location, n (%) | 0.842 | |||
| Left | 186 (54.87) | 18 (51.43) | 168 (55.26) | |
| Right | 42 (12.39) | 4 (11.43) | 38 (12.50) | |
| NA | 111 (32.74) | 13 (37.14) | 98 (32.24) | |
| TNM, n (%) | 0.5788 | |||
| I | 3 (0.88) | 1 (2.86) | 2 (0.66) | |
| II | 9 (2.65) | 0 | 9 (2.96) | |
| III | 25 (7.37) | 3 (8.57) | 22 (7.24) | |
| IV | 229 (67.55) | 24 (68.57) | 205 (67.43) | |
| NA | 73 (21.53) | 7 (20.00) | 66 (21.71) | |
| TMB, n (%) | 0.0004* | |||
| TMB-H | 29 (8.55) | 9 (25.71) | 20 (6.58) | |
| TMB-L | 296 (87.32) | 26 (74.29) | 270 (88.82) | |
| NA | 14 (4.13) | 0 | 14 (4.61) | |
| MSI, n (%) | 0.0003* | |||
| MSS | 302 (89.09) | 28 (80.00) | 280 (92.11) | |
| MSI-H | 19 (5.60) | 7 (20.00) | 12 (3.95) | |
| NA | 18 (5.31) | 0 | 12 (3.95) | |
| BRAF mutation type, n (%) | NA | |||
| Class 1 | – | 17 (48.57) | – | |
| Class 2 | – | 4 (11.43) | – | |
| Class 3 | – | 9 (25.71) | – | |
| Class NA (unknown) | – | 5 (14.29) | – |
*, statistical significance. NA, not available; TNM, Tumor-Node-Metastasis; TMB, tumor mutational burden; TMB-H, high TMB; TMB-L, low TMB; MSI, microsatellite instability; MSS, microsatellite stability; MSI-H, high MSI.
We also conducted a cohort comparison of Chinese and Western populations from Memorial Sloan-Kettering Cancer Center (MSKCC) (22,23). On the one hand, MSKCC datasets focusing on metastasis CRC were selected to match the Chinese cohort TNM (Tumor-Node-Metastasis) staging, and on the other hand, batch datasets could eliminate the bias of artificial selection of data, thus making the analysis results more credible. The somatic mutation data and the clinicopathological information of patients with 125 CRC data were downloaded from the cBioPortal platform (http://www.cbioportal.org).
DNA isolation and targeted capture sequencing
Genome DNA (gDNA) was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue samples and peripheral blood lymphocytes (PBL) using TIANamp Blood DNA Kits and TIANamp Genomic DNA Kits (Tiangen Biotech, China), respectively. DNA quality was controlled using Qubit®2.0 fluorimeter (Life Technologies, USA) and 2100 Bioanalyzer (Agilent, USA). Target DNA capture was performed using the 1,021 panel (Geneplus, China; table available at https://cdn.amegroups.cn/static/public/cco-23-117-1.xlsx), a custom-designed biotinylated oligonucleotide probes (Roche NimbleGen, Madison, WI, USA) covering ~1.4 Mbp coding region of genomic sequence of 1,021 cancer-related genes. Libraries were constructed using the Hieff NGS Ultima DNA Library Prep Kit (Yeasen Biotechnology, Shanghai, China). High throughput sequencing was performed on an Illumina HiSeq 2000/2500 platform (Illumina, San Diego, CA, USA) to generate 101 bp paired-end reads.
NGS analysis
Quality metrics were performed on raw data using NCfilter and aligned to the human genome build GRCh37 using Burrows-Wheeler Aligner (BWA) (24). Picard (http://broadinstitute.github.io/picard/) were used to mark PCR duplicates. Somatic mutation single nucleotide variant (SNV) and Indels were performed using MuTect2 (25) and GATK (26), respectively. Somatic structural variant (SV) mutation was called using NCsv software (Geneplus, China). PBL NGS results were used to filter germline mutation. All candidate somatic mutations were manually reviewed using Integrated Genomics Viewer (IGV) (27) to filter out false positives. The mutated protein coding position and filtered intronic and silent changes were annotated by ANNOVAR software (28). The 1,021-panel has corrected coverage data for GC content and sequencing bias resulting from probe design, which can eliminate bias in mutation analysis.
Tumor mutational burden (TMB) and microsatellite instability (MSI) status analysis
The TMB in the Chinese cohort was defined as the number of non-silent somatic mutations (non-synonymous single nucleotide variation “SNV”, indel, and splice ±2) per mega-base (1 Mb) of coding genomic regions sequenced (1.03 Mb for this 1,021 panel) (29). The TMB in Western cohort from MSKCC adds an additional frameshift variant type. In the present study, the upper quartile of TMB was deemed as high TMB (TMB-H) (22,23). The threshold values of the Chinese cohort and Western cohort were 9 and 11.74 mutations/Mb, respectively. The MSI status of NGS data in Chinese cohort were inferred using MSIsensor (v0.2), which reported the percentage of unstable somatic microsatellites through Chi-square test on predefined microsatellite regions covered by 1,021 panel. Default parameters were used (30). The Western cohort of MSI status was also calculated by MSI sensor (22,31) and the data were download as described above.
Statistical analysis
Analysis of differences among two groups were calculated and presented using either Fisher’s exact test or paired, two-tailed Student’s t-test. Univariate logistic regression was used to analyze the correlation between BRAF mutation status, others mutation and clinical features. Survival curves were estimated by Kaplan-Meier method and differences among designed groups were tested by log-rank test. P values <0.05 was denoted as statistically significant. All the data were analyzed using R statistics package (R version 4.2.1, Austria) or GraphPad Prism version 8 (GraphPad Software, CA, USA).
Mutation analysis and functional enrichment
Co-occurrence mutation analysis was performed using R statistics package, which was used to explore consistency and differences in class 1/2/3/NA BRAF mutation cohort. Also, in this present study, we explored the main biological functions of the identified statistically significant gene via the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. David 6.8 (https://david.ncifcrf.gov/) was used to carried out the GO and KEGG pathway enrichment analysis.
Results
Patient characteristics
Overall, 339 Chinese CRC patients were retrospectively reviewed in Beijing Hospital. Thirty-five (10.32%) patients carried BRAF mutation, with a total of 17 patient tissues (48.57%) were BRAF V600E. All patients were updated according to the 8th edition of the TNM staging criteria. Most patients were diagnosed at stage IV disease (229/339, 67.55%). Besides, patients with primary cancer were the most type (230/339, 67.85%). Regarding primary tumor location, 186 (54.87%), 42 (12.39%), and 111 (32.74%) patients were diagnosed with left-sided, right-sided and NA primary tumor location in total patients. There were 4 (11.43%) right-sided (cecum to transverse colon) and 18 (51.43%) left-sided (splenic flexure to sigmoid colon) in BRAF-mutated population. This was inconsistent with previous literatures (7), which may be related to the sample size, but there was no significant statistical difference (P=0.842). According to our analysis, patients with BRAF mutation had significant association with TMB-H and high MSI (MSI-H) than those with BRAF wild-type (P=0.0004 for TMB, P=0.0003 for MSI). The clinical and pathological features are showed in Table 1.
In this study, 125 Western CRC patients, including 67 men (53.60%) and 58 women (46.40%), were brought into study from the cBioPortal database. Most patients were diagnosed at stage IV disease (72/125, 57.60%). The frequencies of age at diagnosis, gender, sample type, stage (TNM), MSI, TMB, and BRAF mutation type were consistent with the Chinese population. However, the primary tumor location showed significant statistical differences (P<0.0001). Fifty-two (41.60%) and 72 (57.60%) were diagnosed with left-sided and right-sided primary tumor location, respectively (table available at https://cdn.amegroups.cn/static/public/cco-23-117-2.xlsx).
Prevalence of BRAF mutation and co-mutation features
Of the analyzed patients, a total of 39 and 134 BRAF mutation were detected in the Chinese cohort and the Western cohort, respectively. Class 1 BRAF mutation, all of them was p.V600E, were mostly dominant mutation (Figure 1). We first analyzed the types and distribution of BRAF mutations, as well as the differences in the incidence of BRAF mutations in the Chinese and Western cohort and pathway enrichment analysis was also performed. Then, we analyzed the mutation spectrum and con-mutation differences between class 1 BRAF mutation and other types of BRAF mutations (based on classification system) (20), and the KEGG pathway enrichment was also performed. We also compared BRAF mutation and BRAF wild-type populations as described in the analysis above.
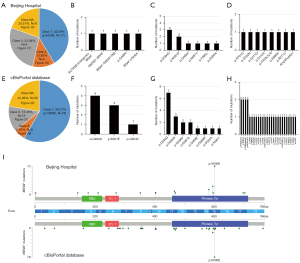
In the Chinese cohort, 35 patients carried 39 BRAF mutation, with 43.59% (17/39), 12.82% (5/39), 23.08% (9/39) and 20.51% (8/39) patients had class 1, 2, 3 and NA BRAF mutations, respectively (Figure 1A). The specific distributions of patients with class 2, 3 and NA were depicted in Figure 1B-1D, respectively. The class 2 BRAF mutation subgroup consisted of four fusion mutation types [ACTR3B (intergenic)-BRAF, MKRN1-BRAF, BRAF-TMEM178B, BRAF-USH2A) and one missense (p.G469V) (table available at https://cdn.amegroups.cn/static/public/cco-23-117-3.xlsx). Of the patients with class 3 BRAF mutation, six missenses were found, including p.D594G [3], p.N581S [2], p.G466V [1], p.N581Y [1], p.F595L [1], p.D594N [1]. Furthermore, 11 BRAF mutations were classified as undefined. Among them, six were missense types (p.R701G, p.D352N, p.D211G, p.D143V, p.E695K, p.R271C), one was frameshift (p.P403Lfs*8) and one was BRAF amplification. Meanwhile, four patients (11.43%, 4/35) had compound BRAF mutation. All of them belong to non-class 1 BRAF mutations (table available at https://cdn.amegroups.cn/static/public/cco-23-117-4.xlsx).
In Western cohort, the distribution of patients with class 1, 2, 3 and NA were 58.21% (78/134), 5.97% (8/134), 13.43% (18/134) and 22.39% (30/134), respectively (Figure 1E). Only three missenses were found in class 2 BRAF mutation, including p.G469A [4], p.K601E [3], p.G464V [1], which may be due to the lack of analysis of SV and copy number variation (CNV) in this study (Figure 1F). Among the class 3 BRAF mutations (Figure 1G), the Western population and the Chinese population showed a high mutation consistency, manifested as a relatively higher mutation frequency of p.D594G and p.N581S. Moreover, 21 missense, one nonsense and eight frameshifts were defined as class NA subgroup, among them manifested the characteristics of wide and irregular distribution (Figure 1H, table available at https://cdn.amegroups.cn/static/public/cco-23-117-3.xlsx).
Next, we investigated the correlation between BRAF mutation and clinicopathological features. In the Chinese cohort, class 1 BRAF mutation was more likely to occur in elder (41.18% vs. 22.22%, P=0.18) and female (58.82% vs. 33.33%, P=0.18), although no statistical difference was presented. Other clinicopathological features, including sample type, primary tumor location, TMB and MSI were not significantly associated with BRAF mutation types (Table 2).
Table 2
| Characteristics | Class 1 (n=17) | Non-class 1 (n=18) | P value |
|---|---|---|---|
| Age, n (%) | 0.18 | ||
| Young | 1 (5.88) | 5 (27.78) | |
| Intermediate | 8 (47.06) | 8 (44.44) | |
| Elder | 7 (41.18) | 4 (22.22) | |
| NA | 1 (5.88) | 1 (5.56) | |
| Gender, n (%) | 0.18 | ||
| Male | 7 (41.18) | 12 (66.67) | |
| Female | 10 (58.82) | 6 (33.33) | |
| Sample type, n (%) | >0.99 | ||
| Primary | 11 (64.71) | 15 (83.33) | |
| Metastasis | 2 (11.76) | 2 (11.11) | |
| NA | 4 (23.53) | 1 (5.56) | |
| Primary tumor location, n (%) | 0.112 | ||
| Left | 9 (52.94) | 9 (50.00) | |
| Right | 4 (23.53) | 0 | |
| NA | 4 (23.53) | 9 (50.00) | |
| MSI, n (%) | >0.99 | ||
| MSI-H | 3 (17.65) | 4 (22.22) | |
| MSS | 14 (82.35) | 14 (77.78) | |
| TMB, n (%) | 0.443 | ||
| TMB-H | 3 (17.65) | 6 (33.33) | |
| TMB-L | 14 (82.35) | 12 (66.67) | |
NA, not available; MSI, microsatellite instability; MSI-H, high MSI; MSS, microsatellite stability; TMB, tumor mutational burden; TMB-H, high TMB; TMB-L, low TMB.
We further analyzed the Western cohort and the data were shown in Table 3. Consistent with the Chinese data, class 1 BRAF mutation was more common in elderly patients (29.49% vs. 14.89%), non-class 1 BRAF mutations were more likely to occur in younger patients (24.36% vs. 42.55%). We compared the distribution of different BRAF mutation types between the genders. The results showed that class 1 were more likely to occur in female than non-class 1 mutations (53.85% vs. 34.04%, P<0.05). There is no significant difference between class 2 vs. class 3, class 1 vs. class 2, and class 1 vs. class 3 (28.57% vs. 41.18%, P=0.67; 53.85% vs. 28.57%, P=0.25; 53.85% vs. 41.18%, P=0.43). When sample type distribution was analyzed, class 1 exhibited primary predominance in both class 1 vs. others (67.95% vs. 46.81%, P<0.05) and class 1 vs. class 3 (67.95% vs. 41.18%, P<0.05). To study primary tumor location, we found right-sided CRC was more likely to occur in class 1 (class 1 vs. others: 69.23% vs. 38.30%, P<0.05; class 1 vs. class 2: 69.23% vs. 28.57%, P<0.05; class 1 vs. class 3: 69.23% vs. 29.41%, P<0.05), which was inconsistent with the data analyzed above (Table 2). Meanwhile, we examined the relationship between genomic markers—TMB, MSI and BRAF mutation types. The data showed that the proportion of TMB-H (57.69% vs. 11.76%, P<0.001) and MSI-H (28.21% vs. 0%, P<0.05) in BRAF class 1 mutations was significantly higher than that in BRAF class 3 mutations.
Table 3
| Characteristics | Class 1 (n=78) | Others (n=47) | P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total (n=47) | Class 2 (n=7) | Class 3 (n=17) | NA (n=23) | Class 1 vs. others | Class 2 vs. class 3 | Class 1 vs. class 2 | Class 1 vs. class 3 | |||
| Age, n (%) | 0.055 | 0.9378 | 0.499 | 0.5087 | ||||||
| Young | 19 (24.36) | 20 (42.55) | 3 (42.86) | 6 (35.29) | 11 (47.83) | |||||
| Intermediate | 36 (46.15) | 20 (42.55) | 3 (42.86) | 8 (47.06) | 9 (39.13) | |||||
| Elder | 23 (29.49) | 7 (14.89) | 1 (14.29) | 3 (17.65) | 3 (13.04) | |||||
| Gender, n (%) | 0.0417* | 0.6687 | 0.2547 | 0.4257 | ||||||
| Male | 36 (46.15) | 31 (65.96) | 5 (71.43) | 10 (58.82) | 16 (69.57) | |||||
| Female | 42 (53.85) | 16 (34.04) | 2 (28.57) | 7 (41.18) | 7 (30.43) | |||||
| Sample type, n (%) | 0.0184* | >0.99 | 0.1941 | 0.0237* | ||||||
| Primary | 53 (67.95) | 22 (46.81) | 3 (42.86) | 7 (41.18) | 12 (52.17) | |||||
| Metastasis | 21 (26.92) | 23 (48.94) | 4 (57.14) | 10 (58.82) | 9 (39.13) | |||||
| NA | 4 (5.13) | 2 (4.26) | 0 | 0 | 2 (8.70) | |||||
| Primary tumor location, n (%) | 0.0014* | >0.99 | 0.0428* | 0.0088* | ||||||
| Left | 24 (30.77) | 28 (59.57) | 5 (71.43) | 11 (64.71) | 12 (52.17) | |||||
| Right | 54 (69.23) | 18 (38.30) | 2 (28.57) | 5 (29.41) | 11 (47.83) | |||||
| NA | 0 | 1 (2.13) | 0 | 1 (5.88) | 0 | |||||
| MSI, n (%) | 0.0642 | 0.2105 | >0.99 | 0.008* | ||||||
| MSS | 41 (52.56) | 31 (65.96) | 3 (42.86) | 15 (88.24) | 13 (56.52) | |||||
| MSI-H | 22 (28.21) | 6 (12.77) | 1 (14.29) | 0 | 5 (21.74) | |||||
| NA | 15 (19.23) | 10 (21.28) | 3 (42.86) | 2 (11.76) | 5 (21.74) | |||||
| TMB, n (%) | 0.139 | 0.1265 | 0.6937 | 0.0009* | ||||||
| TMB-H | 45 (57.69) | 20 (42.55) | 3 (42.86) | 2 (11.76) | 15 (65.22) | |||||
| TMB-L | 33 (42.31) | 27 (57.45) | 4 (57.14) | 15 (88.24) | 8 (34.78) | |||||
*, statistical significance. NA, not available; MSI, microsatellite instability; MSI-H, high MSI; MSS, microsatellite stability; TMB, tumor mutational burden; TMB-H, high TMB; TMB-L, low TMB.
Concurrent oncogenic mutations
We further analyzed the distribution of concurrent oncogenic mutations between the Chinese cohort and Western cohort. A total of 339 Chinese patients were tested by 1,021-gene panel NGS, 125 Western patients were tested by MSK-IMPACT (table available at https://cdn.amegroups.cn/static/public/cco-23-117-5.xlsx).
The rate of concomitant mutation of class 1/2/3/NA BRAF mutations in patients from the two cohorts are given in Figure 2. Collectively, the top 10 concurrent mutation frequencies of class 1 BRAF mutation in the Chinese population were in TP53 (tumor protein p53) (65%), PTEN (phosphatase and tensin homolog) (35%), RNF43 (29%), MLL2 (24%), MLL3 (24%), NOTCH1 (24%), PIK3CA (24%), SMAD4 (24%), TGFBR2 (24%) and APC (APC regulator of WNT signaling pathway) (18%). It was found that APC (67%) was the highest mutation frequency in top 10 non-class 1 BRAF mutations, followed by TP53 (67%), KRAS (56%), LRP1B (44%), FBXW7 (39%), AXIN2 (28%), FGFR1 (fibroblast growth factor receptor 1) (28%), NOTCH1 (28%), PIK3CA (28%) and PPP2R1A (28%).
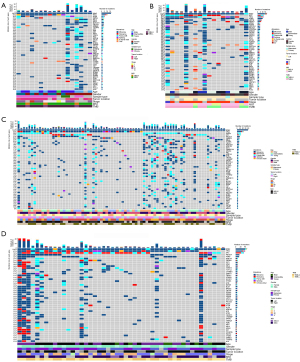
Class 1 BRAF mutation has a lower concomitant mutation frequency, and also tends to be accompanied by co-mutation of passenger genes in the Chinese population (Figure 2A,2B). Identical results were obtained from the Western population (Figure 2C,2D). In addition, we compared BRAF mutation types and wild-type in the Chinese population. The top 10 concomitant mutations of BRAF mutation in the Chinese population were in TP53 (66%), APC (43%), LRP1B (31%), KRAS (29%), FBXW7 (26%), NOTCH1 (26%), PIK3CA (26%), FAT2 (20%), MLL2 (20%), and MLL3 (20%), while those of BRAF wild-type (table available at https://cdn.amegroups.cn/static/public/cco-23-117-6.xlsx) were in TP53 (77%), APC (67%), KRAS (49%), PIK3CA (18%), SMAD4 (18%), TCF7L2 (18%), FBXW7 (17%), MYC (17%), LRP1B (16%), and PTEN (11%). We can find that BRAF wild-type is associated with higher frequencies of concomitant mutation, mainly manifested in the tumor suppressor genes TP53 and APC, as well as tumor driver gene KRAS in the Chinese cohort. The above two findings did not show significant differences (Figure S1). The results suggest that class 1/non-class 1 BRAF mutations and BRAF wild-type have significant differences in carcinogenicity.
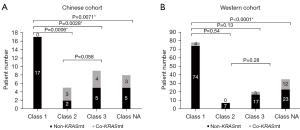
Meanwhile, we compared the co-occurrence and mutual exclusion of core driver gene mutations in the two cohorts (Table 4). KRAS was significantly enriched in the non-class 1 BRAF mutations, indicating that KRAS and class 1 BRAF mutation are mutually exclusive in the Chinese cohort (P=0.0003), and Western cohort (P=0.0001). None of the 35 patients with class 1 BRAF mutation had co-KRAS mutation. However, three patients with class 2, four patients with class 3 and three patients with class NA BRAF mutation had concurrent oncogenic KRAS mutation (most of the sites were p.A146X, and no p.G12C appeared) (P<0.001) in the Chinese cohort. At the same time, we analyzed the Western cohort data, and found four class 1 BRAF-mutated patients co-occurred with KRAS (p.G12A, p.G13D, p.I171Nfs*14, p.G12D). The numbers of class 2, class 3 and class NA BRAF-mutated patients with co-KRAS mutation were 0, 3 and 12, respectively. In addition, we found that there was no class 2 BRAF mutations in the Western cohort, which may be related to the absence of fusion mutation in the MSKCC, because there were three class 2 co-mutation patients in the Chinese cohort, all of which were BRAF fusion with co-KRAS mutation. The result is shown in Figure 3. Also, we found that co-APC mutation was significantly enriched in the non-class 1 BRAF mutations (P=0.0059 in the Chinese cohort, P<0.0001 in the Western cohort). Since the APC gene is a typical diagnostic marker for CRC, it has a significantly longer tumor formation period, which suggests that patients with non-class 1 BRAF mutations have a longer tumor formation period and relatively lighter carcinogenesis than class 1 patients. Considering the limited race characteristics, differences between the two cohorts may have occurred. Co-PTEN mutation and co-FGFR1 mutation were significantly enriched in the class 1 BRAF-mutated and non-class 1 BRAF-mutated Chinese population, respectively, while no statistical difference was found in the Western cohort. In contrast, co-RNF43 mutation and non-TP53 mutation were significantly enriched in the class 1 BRAF-mutated Western population, whereas in the Chinese cohort, no statistical difference was found.
Table 4
| Cohort | Characteristics | Class 1 | Non-Class 1 | P value | 95% CI |
|---|---|---|---|---|---|
| Beijing Hospital | Co-KRASmt | 0 | 10 | 0.0003 | 0.000 to 0.2417 |
| Non-KRASmt | 17 | 8 | |||
| cBioPortal database | Co-KRASmt | 4 | 15 | 0.0001 | 0.03990 to 0.3752 |
| Non-KRASmt | 74 | 32 | |||
| Beijing Hospital | Co-APCmt | 3 | 12 | 0.0059 | 0.02696 to 0.5510 |
| Non-APCmt | 14 | 6 | |||
| cBioPortal database | Co-APCmt | 27 | 35 | <0.0001 | 0.08421 to 0.3965 |
| Non-APCmt | 51 | 12 | |||
| Beijing Hospital | Co-PTENmt | 6 | 1 | 0.0408 | 1.072 to 111.5 |
| Non-PTENmt | 11 | 17 | |||
| cBioPortal database | Co-PTENmt | 10 | 7 | 0.791 | 0.3179 to 2.205 |
| Non-PTENmt | 68 | 40 | |||
| Beijing Hospital | Co-FGFR1mt | 0 | 5 | 0.0455 | 0.000 to 0.4643 |
| Non-FGFR1mt | 17 | 13 | |||
| cBioPortal database | Co-FGFR1mt | 3 | 2 | >0.9999 | 0.1784 to 5.226 |
| Non-FGFR1mt | 75 | 45 | |||
| Beijing Hospital | Co-FGFR1mt | 5 | 2 | 0.2285 | 0.05445 to 1.971 |
| Non-FGFR1mt | 12 | 16 | |||
| cBioPortal database | Co-FGFR1mt | 28 | 4 | 0.0002 | 2.542 to 26.78 |
| Non-FGFR1mt | 50 | 43 | |||
| Beijing Hospital | Co-TP53mt | 11 | 12 | >0.9999 | 0.2496 to 3.354 |
| Non-TP53mt | 6 | 6 | |||
| cBioPortal database | Co-TP53mt | 47 | 41 | 0.0013 | 0.08688 to 0.5595 |
| Non-TP53mt | 31 | 6 | |||
| Beijing Hospital | Co-FBXW7mt | 2 | 7 | 0.1212 | 0.03991 to 1.078 |
| Non-FBXW7mt | 15 | 11 | |||
| cBioPortal database | Co-FBXW7mt | 15 | 8 | 0.8158 | 0.4744 to 2.975 |
| Non-FBXW7mt | 63 | 39 | |||
| Beijing Hospital | Co-LRP1Bmt | 3 | 8 | 0.1464 | 0.06666 to 1.221 |
| Non-LRP1Bmt | 14 | 10 | |||
| cBioPortal database | Co-LRP1Bmt | NA | NA | NA | NA |
| Non-LRP1Bmt | NA | NA | |||
| Beijing Hospital | Co-AXIN2mt | 1 | 5 | 0.1774 | 0.01311 to 1.217 |
| Non-AXIN2mt | 16 | 13 | |||
| cBioPortal database | Co-AXIN2mt | 8 | 3 | 0.5334 | 0.4699 to 6.072 |
| Non-AXIN2mt | 70 | 44 | |||
| Beijing Hospital | Co-PPP2R1Amt | 1 | 5 | 0.1774 | 0.01311 to 1.217 |
| Non-PPP2R1Amt | 16 | 13 | |||
| cBioPortal database | Co-PPP2R1Amt | NA | NA | NA | NA |
| Non-PPP2R1Amt | NA | NA |
Mut, mutation; CI, confidence interval; NA, not available.
KEGG pathway and GO enrichment
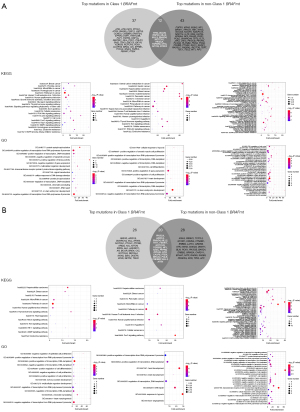
We analyzed the enrichment results of signature mutations of class 1 and non-class 1 populations. We first counted the unique mutations between class 1 BRAF mutation and non-class 1 BRAF mutations, and then used the Fisher test to count the incidence of genes that coexist in the two groups. Genes with P<0.05 and odds ratio (OR) >1 were incorporated into non-class 1 unique mutation set, while genes with P<0.05 and OR <1 were incorporated into class 1 BRAF-mutated unique mutation set.
The KEGG pathway enrichment results showed that class 1 BRAF mutation was enriched with fewer proto-cancer signaling pathways, which further proved that class 1 BRAF mutation has stronger tumorigenicity. GO enrichment results showed that the most common pathway in class 1 BRAF mutation was GO:0000122/0045944 (type II RNA polymerase promoter transcriptional regulator), while the pathway in the non-class 1 BRAF mutations included GO:0008285 (cell proliferation regulation), GO:0010628 (regulation of gene expression), GO:0051726 (regulation of cell cycle), etc., which prove that class 1 BRAF mutation has stronger tumorigenicity (Figure 4). Meanwhile, we explored the difference in KEGG pathway enrichment between BRAF wild type and BRAF mutations, and the results showed that compared with BRAF wild type, BRAF mutations have stronger carcinogenicity, which is consistent with the enrichment pathway analyzed above [GO:0000122/0045944 (type II RNA polymerase promoter transcriptional regulator) pathway] (Figure S2).
Survival outcomes
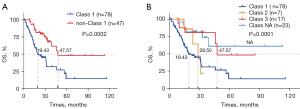
We also analyzed survival outcomes based on BRAF mutation types in 125 patients with evaluable stage I–IV CRC (Figure 5). First, we compared the survival outcomes of 78 patients with class 1 BRAF mutation and 47 patients with non-class 1 BRAF mutations. Kaplan-Meier and log-rank analysis showed that patients with non-class 1 BRAF mutations had longer OS (P=0.0002), with median OS of 47.57 vs. 19.43 months, respectively. Second, we divided BRAF mutations into four categories: class 1 (n=78)/2 (n=7)/3 (n=17)/NA (n=23). Kaplan-Meier and log-rank analysis showed that the median OS of the four types of BRAF population was 19.43 vs. 28.50 vs. 47.57 months vs. not reached (P=0.0001).
Discussion
Our study tried to compare the differences in concomitant mutational patterns between the Chinese and Western populations with CRC and their correlation with clinicopathological features. We found that class 1 BRAF mutation was more common in elderly and female patients in both Chinese and Western populations, while non-class 1 BRAF mutations were more likely to occur in younger patients in the Western population. Additionally, we found that class 1 BRAF mutation was more likely to be accompanied by passenger gene mutations, and rich in oncogenic signaling pathways compared to non-class 1 BRAF mutations in the Chinese population. This means that BRAF V600E could be the main important driver mutation. Furthermore, our results showed that TMB-H and MSI-H were significantly associated with class 1 BRAF mutation in the Western population.
Recognizing the co-occurrence of BRAF V600E with other gene mutations in CRC patients is important because that may affect treatment outcomes. Several studies have shown that potential biomarkers including high BRAF allele scores (≥2%) (17), RNF43 mutation (18), consensus molecular subtypes (CMS) (19), and POLD1/POLE mutation (32) might bring clinical benefits from different treatment modalities. In our study, we found co-mutation features of BRAF mutations with other genes such as KRAS, APC, PTEN, and TP53. Therefore, identifying and evaluating the co-mutation status of BRAF and other genes in CRC patients will help develop personalized treatment strategies. For example, patients with BRAF and KRAS co-mutations have shown to have a poorer prognosis compared to patients with BRAF or KRAS mutations alone (33). In addition, co-mutations of BRAF have been associated with resistance to chemotherapy and poor survival outcomes and may benefit from immunotherapy (34,35). The limitation of this study is that it is retrospective and it is not possible to obtain neat treatment and PFS/OS data. Therefore, this study only conducted prognostic correlation analysis for BRAF mutation types. Furtherly, prospective clinical trials are needed to explore therapeutic benefits.
Based on the results of this study, we observed a significant association between BRAF mutations and TMB-H and MSI-H. However, TMB-H may be associated with MSI-H rates in patients with BRAF-mutated tumors, which is consistent with published literature (36) showing that a majority of MSI-H samples are also TMB-H (83%), and 97% had TMB ≥10 mutations/Mb. These findings suggest that immunotherapy may be an effective treatment option for CRC patients with BRAF V600E mutations. Now we all know targeted therapy may also be a viable treatment option for CRC patients with BRAF V600E. Several targeted therapies, such as vemurafenib and dabrafenib, have been approved by the Food and Drug Administration (FDA) for the treatment of BRAF V600E melanoma and second-line for mCRC. Recently, in addition to targeted therapy, several clinical trials have shown that immune checkpoint inhibitors, such as pembrolizumab and nivolumab, are effective in treating BRAF V600E CRC (37,38). One of the most promising agents is pembrolizumab, which has been approved by the FDA for the treatment of advanced CRC with specific genetic mutations, including BRAF V600E (39). Therefore, combining immunotherapy with targeted therapy may be a better choice for treating CRC patients with BRAF V600E in China. However, further studies are needed to validate these findings and to determine the optimal treatment strategy for CRC patients with BRAF V600E.
Conclusions
In conclusion, our study showed that immunotherapy and targeted therapy may be effective treatment options for CRC patients with BRAF V600E, particularly when combined with targeted therapies. However, the co-occurrence of BRAF with other gene mutations in CRC patients may affect treatment outcomes, and personalized treatment strategies are needed. Further studies are warranted to validate these findings and to identify optimal treatment regimens for CRC patients with BRAF V600E.
Acknowledgments
A part of the manuscript has been presented in ASCO 2023 annual meeting (doi: 10.1200/JCO.2023.41.16_suppl.e15634). A main conclusion of the manuscript has not been published previously. We agree to transfer copyright and assign all rights to the manuscript (and any revisions or versions of it) solely to the journal, to print, publish, distribute, and sell the article in all languages and media internationally.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://cco.amegroups.com/article/view/10.21037/cco-23-117/rc
Data Sharing Statement: Available at https://cco.amegroups.com/article/view/10.21037/cco-23-117/dss
Peer Review File: Available at https://cco.amegroups.com/article/view/10.21037/cco-23-117/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://cco.amegroups.com/article/view/10.21037/cco-23-117/coif). Z.L. is an employee of Geneplus-Beijing (Beijing, China). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Hospital (No. 2023BJYYEC-103-01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. Erratum in: CA Cancer J Clin 2021;71:359. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Abushukair HM, Zaitoun SM, Saeed A. Insight on BRAF(V600E) mutated colorectal cancer immune microenvironment. World J Gastrointest Oncol 2022;14:1213-5. [Crossref] [PubMed]
- Huang Y, Jia W, Zhao G, et al. Analysis of driver gene mutations in colorectal cancer by using next-generation sequencing. Chinese Journal of Geriatrics 2021;12:646-9.
- Tabernero J, Ros J, Élez E. The Evolving Treatment Landscape in BRAF-V600E-Mutated Metastatic Colorectal Cancer. Am Soc Clin Oncol Educ Book 2022;42:1-10. [Crossref] [PubMed]
- Guan WL, Qiu MZ, He CY, et al. Clinicopathologic Features and Prognosis of BRAF Mutated Colorectal Cancer Patients. Front Oncol 2020;10:563407. [Crossref] [PubMed]
- Morkel M, Riemer P, Bläker H, et al. Similar but different: distinct roles for KRAS and BRAF oncogenes in colorectal cancer development and therapy resistance. Oncotarget 2015;6:20785-800. [Crossref] [PubMed]
- Chen G, McQuade JL, Panka DJ, et al. Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib Combination Treatment for BRAF Inhibitor-Refractory Metastatic Melanoma: A Phase 2 Clinical Trial. JAMA Oncol 2016;2:1056-64. [Crossref] [PubMed]
- Planchard D, Groen HJ, Kim TM, et al. Interim results of a phase II study of the BRAF inhibitor (BRAFi) dabrafenib (D) in combination with the MEK inhibitor trametinib (T) in patients (pts) with BRAF V600E mutated (mut) metastatic non-small cell lung cancer (NSCLC). J Clin Oncol 2015;33:8006. [Crossref]
- Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012;366:707-14. [Crossref] [PubMed]
- Corcoran RB, Atreya CE, Falchook GS, et al. Combined BRAF and MEK Inhibition With Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J Clin Oncol 2015;33:4023-31. [Crossref] [PubMed]
- Tabernero J, Grothey A, Van Cutsem E, et al. Encorafenib Plus Cetuximab as a New Standard of Care for Previously Treated BRAF V600E-Mutant Metastatic Colorectal Cancer: Updated Survival Results and Subgroup Analyses from the BEACON Study. J Clin Oncol 2021;39:273-84. [Crossref] [PubMed]
- Kopetz S, Guthrie KA, Morris VK, et al. Randomized Trial of Irinotecan and Cetuximab With or Without Vemurafenib in BRAF-Mutant Metastatic Colorectal Cancer (SWOG S1406). J Clin Oncol 2021;39:285-94. [Crossref] [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [Crossref] [PubMed]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227-35. [Crossref] [PubMed]
- Ros J, Matito J, Villacampa G, et al. Plasmatic BRAF-V600E allele fraction as a prognostic factor in metastatic colorectal cancer treated with BRAF combinatorial treatments. Ann Oncol 2023;34:543-52. [Crossref] [PubMed]
- Elez E, Ros J, Fernández J, et al. RNF43 mutations predict response to anti-BRAF/EGFR combinatory therapies in BRAF(V600E) metastatic colorectal cancer. Nat Med 2022;28:2162-70. [Crossref] [PubMed]
- Kopetz S, Murphy D, Pu J, et al. Molecular correlates of clinical benefit in previously treated patients (pts) with BRAF V600E-mutant metastatic colorectal cancer (mCRC) from the BEACON study. J Clin Oncol 2021;39:3513. [Crossref]
- Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234-8. [Crossref] [PubMed]
- Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183-99. [Crossref] [PubMed]
- Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell 2018;33:125-136.e3. [Crossref] [PubMed]
- Mondaca S, Walch H, Nandakumar S, et al. Specific Mutations in APC, but Not Alterations in DNA Damage Response, Associate With Outcomes of Patients With Metastatic Colorectal Cancer. Gastroenterology 2020;159:1975-1978.e4. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213-9. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24-6. [Crossref] [PubMed]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [Crossref] [PubMed]
- Peng W, Li B, Li J, et al. Clinical and genomic features of Chinese lung cancer patients with germline mutations. Nat Commun 2022;13:1268. [Crossref] [PubMed]
- Wang J, Yi Y, Xiao Y, et al. Prevalence of recurrent oncogenic fusion in mismatch repair-deficient colorectal carcinoma with hypermethylated MLH1 and wild-type BRAF and KRAS. Mod Pathol 2019;32:1053-64. [Crossref] [PubMed]
- Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015-6. [Crossref] [PubMed]
- Garmezy B, Gheeya JS, Thein KZ, et al. Correlation of pathogenic POLE mutations with clinical benefit to immune checkpoint inhibitor therapy. J Clin Oncol 2020;38:3008. [Crossref]
- Won DD, Lee JI, Lee IK, et al. The prognostic significance of KRAS and BRAF mutation status in Korean colorectal cancer patients. BMC Cancer 2017;17:403. [Crossref] [PubMed]
- Taieb J, Lapeyre-Prost A, Laurent Puig P, et al. Exploring the best treatment options for BRAF-mutant metastatic colon cancer. Br J Cancer 2019;121:434-42. [Crossref] [PubMed]
- Park R, Lopes L, Lee S, et al. The prognostic and predictive impact of BRAF mutations in deficient mismatch repair/microsatellite instability-high colorectal cancer: systematic review/meta-analysis. Future Oncol 2021;17:4221-31. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Ros J, Saoudi N, Baraibar I, et al. Encorafenib plus cetuximab for the treatment of BRAF-V600E-mutated metastatic colorectal cancer. Therap Adv Gastroenterol 2022;15:17562848221110644. [Crossref] [PubMed]
- Morris VK, Parseghian CM, Escano M, et al. Phase I/II trial of encorafenib, cetuximab, and nivolumab in patients with microsatellite stable, BRAFV600E metastatic colorectal cancer. J Clin Oncol 2022;40:12. [Crossref]
- Casak SJ, Marcus L, Fashoyin-Aje L, et al. FDA Approval Summary: Pembrolizumab for the First-line Treatment of Patients with MSI-H/dMMR Advanced Unresectable or Metastatic Colorectal Carcinoma. Clin Cancer Res 2021;27:4680-4. [Crossref] [PubMed]


