MicroRNAs in nasopharyngeal carcinoma
Introduction
MicroRNAs (miRNAs; miRs) have a fundamental role in cancer initiation, progression, and treatment response. Recent genomic studies have identified deregulation of several miRNAs in nasopharyngeal carcinoma (NPC), and have identified potentially clinically relevant prognostic miRNA signatures (1-3). Based on a number of reports that have described the presence of tumour-specific biomarkers circulating in the plasma of patients for many tumour types (4,5); tumour-specific miRNA signatures might prove to be highly useful as early, non-invasive tools for diagnosis and prognosis. Furthermore, in addition to a number of miRNAs encoded in the human genome that are determined to be deregulated in NPC, expression of miRNAs encoded by the Epstein-Barr virus (EBV), known to be the most common causal agent in NPC (6,7), have also been detected in these tumours. EBV-associated miRNAs might well be functioning as drivers of NPC tumorigenesis and progression. In addition to their enormous potential as putative biomarkers, exploring the role of miRNAs in NPC pathogenesis will likely inform important insights into NPC biology, including regulation of proliferation, migration, invasion, and apoptosis, as well as resistance to chemotherapy and radiation therapy. In this article, we will review the functional role of miRNAs in NPC pathogenesis, and examine their potential for use as prognostic and predictive biomarkers.
MicroRNAs
miRNAs are a family of endogenous, non-coding RNAs of approximately 22 nucleotides in length (8). miRNAs have been implicated as both oncogenes and tumour suppressors (9), importantly influencing tumour initiation, progression, and response to treatment. Functionally, miRNAs predominantly act by directly binding to specific sequences in target mRNA transcripts to repress protein translation (8). Alternative mechanisms of regulation also exist, such as positive regulation of mRNA targets, or immediate degradation of mRNA transcripts (10-12). A single miRNA may target thousands of downstream targets based on in silico prediction algorithms (13), rendering them as powerful mediators of transcriptional regulation, as well as highlighting their potential as therapeutic targets.
Biogenesis and processing of microRNAs
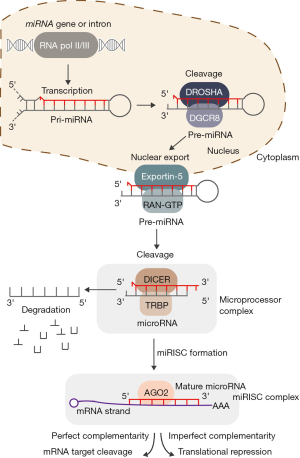
The majority of miRNAs are encoded within intergenic regions and produced from distinct loci, although approximately 30% of miRNAs are encoded within intronic regions of protein coding genes (14). During canonical miRNA biogenesis (see Figure 1), transcription of miRNAs occurs primarily via RNA polymerase II (Pol II), either from a miRNA specific promoter or from the promoter of the gene in which they reside, though RNA polymerase III (Pol III) is also responsible for transcription of a subset of miRNAs (15,16). First, miRNAs are transcribed into primary miRNAs (pri-miRNA), which exist as stem-loop structures and vary from hundreds to thousands of base-pairs in length (17). Pri-miRNAs are then cleaved by the Microprocessor complex, composed of DROSHA, a double-stranded RNase III, and DiGeorge syndrome critical region 8 (DGCR8), a double-stranded RNA binding protein (18,19). A pri-miRNA sequence can produce an individual miRNA or encode clusters of two or three miRNAs. The Microprocessor cleaves the pri-miRNA into a ~60–70 nucleotide stem-loop structure called the precursor miRNA (pre-miRNA), which is then exported from the nucleus to the cytoplasm by Exportin-5 and ras-related nuclear protein guanosine triphosphate (RAN-GTP) for further processing by the RNase III Dicer and TAR RNA-binding protein (TRBP) (17). The mature miRNA is preferentially derived from the guiding strand of the pre-miRNA duplex, and is subsequently bound by the miRNA-induced silencing complex (miRISC). The miRISC complex mediates sequence specific binding to target mRNA transcripts, subsequently leading to either mRNA degradation or translational inhibition (19,20).
MicroRNAs in cancer
A number of important biological processes are regulated by miRNAs through their ability to regulate mRNA levels post-transcriptionally via direct binding and target cleavage, or translational repression (8,13). A single miRNA regulates the expression of a large cohort of mRNAs, often regulating many functionally related pathways to simultaneously control a broad cellular function such as differentiation, proliferation, or apoptosis. Thus, abnormal expression of miRNAs may lead to malignant transformation (21-25), and the association between miRNA deregulation and cancer has been reported in almost all human malignancies, with numerous miRNAs characterized as key oncogenes or tumour suppressors. Additionally, miRNAs are frequently located at fragile sites of the genome (26), highlighting their susceptibility to mutational events. Furthermore, comprehensive expression profiling of miRNAs in cancer has uncovered the potential for miRNA expression signatures to predict response to treatment and patient outcome (4,27,28).
MicroRNAs in nasopharyngeal carcinoma
The majority of NPC cases (75–90%) are diagnosed once the disease has reached an advanced stage (29), which attributes to the high level of metastasis, high risk of recurrence, and poor outcomes observed for NPC patients. Furthermore, nearly all cases of NPC are associated with EBV (6,7) despite its rare presence in normal adjacent epithelial tissue (30), highlighting the potential of EBV as a biomarker for NPC diagnosis. Importantly, EBV expresses few viral proteins in NPC (31); however, several EBV-encoded miRNAs are highly expressed, suggesting the pathogenic mechanisms of NPC are not only regulated by EBV-encoded proteins but underscore the potential importance of EBV-associated miRNAs in NPC. Thus, a thorough understanding of the expression patterns and function of EBV-encoded miRNAs, particularly with respect to their utility as early diagnostic biomarkers and as prognostic biomarkers for aggressive disease, could be potentially relevant and important for NPC detection and disease management.
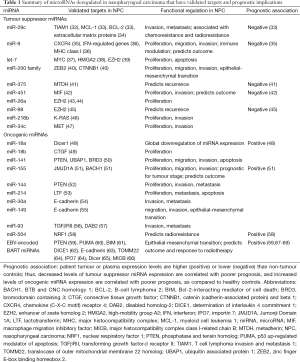
Though thousands of miRNAs have been discovered within the past decade, with advancements in DNA sequencing, we anticipate that a number of additional NPC associated miRNAs will emerge. Circulating miRNAs associated with NPC are of particular interest, as they present an exploitable tool for early detection, diagnosis and staging, as well as prognosis and treatment outcome prediction. Several global profiling studies have recently demonstrated that altered miRNA expression occurs in a spectrum of head and neck cancers. In NPC specifically, under-expression of the tumour suppressor miRNAs, including miR-29c, miR-9, let-7 family, miR-200 family, and overexpression of oncogenic miRNAs, such as miR-18a/b, miR-141, miR-155, miR-214, have been observed, as described in the following section. A summary of miRNAs with validated targets that are deregulated in NPC is shown in Table 1.
Full table
Tumour suppressor microRNAs
Expression of miR-29c is significantly diminished in NPC as compared to normal nasopharyngeal epithelium, and miR-29c has been shown to directly inhibit numerous targets involved in extracellular matrix synthesis and function (34). Furthermore, miR-29c also mediates its tumour suppressive effects through inhibition of the T cell lymphoma invasion and metastasis 1 (TIAM1) gene, thereby promoting migration and invasion of NPC cells (32). Importantly, reduced expression levels of miR-29c have been associated with chemoresistance and radioresistance in primary NPC as well as in vitro and in vivo cell models, mediated by targeting anti-apoptotic proteins myeloid cell leukemia 1 (MCL-1) and B-cell lymphoma 2 (BCL-2) (33).
One of the most commonly implicated miRNAs in NPC pathogenesis is miR-9. In addition to NPC, this miRNA has been implicated in numerous other malignancies, and its tumour suppressive functions have been thoroughly described. miR-9 is a highly conserved miRNA (70) and appears to function in the regulation of numerous essential cellular processes, mediating proliferation, apoptosis, invasion, metastasis, angiogenesis and epithelial-mesenchymal transition (EMT) (71-73). Low levels of miR-9 expression in NPC are associated with more aggressive phenotypes and poorer survival. miR-9 has been shown to function as a tumour suppressor in NPC by targeting chemokine (C-X-C motif) receptor 4 (CXCR4) to inhibit cell proliferation, migration and invasion (35). miR-9 also regulates the innate immune response in NPC through regulation of multiple genes that are induced by interferon, as well as MHC class I molecules (36). Importantly, the ability of miR-9 to function as an individual prognostic biomarker for NPC metastasis has also been demonstrated, whereby miR-9 expression is correlated with reduced proliferation, migration and invasion in NPC cells, and low-levels of miR-9 expression are correlated with advanced tumour stage (74).
The tumour suppressive role of the let-7 family of miRNAs has been well documented in a multitude of tumour types as well. In NPC, expression of the let-7 family of miRNAs is globally reduced (37,75) and these miRNAs have been shown to directly regulate key oncogenic targets in NPC, such as MYC (37), high-mobility group A2 (HMGA2) (38), and enhancer of zeste homolog 2 (EZH2) (39). Thus, reduced let-7 family miRNA expression in NPC leads to increased cellular proliferation and reduced apoptosis.
The miR-200 family is also downregulated in NPC, resulting in increased cell growth, migration and invasion of NPC cells as a result of repression of the putative miR-200 targets zinc finger E-box binding homeobox 2 (ZEB2) and catenin (cadherin-associated protein) and beta 1 (CTNNB1), resulting in increased NPC cell growth, migration and invasion (40). Furthermore, decreased miR-200a expression is associated with inducing epithelial-mesenchymal transition, with miR-200a over-expression conversely linked with a more epithelial-like state in NPC cells, a process that occurs via the regulation of ZEB2 and β-catenin signaling by miR-200a (40).
Numerous other miRNAs have been implicated as tumour suppressors in NPC, including miR-375, miR-451, miR-26a, miR-98, miR-216b, and miR-34c. miR-375 has been reported as a potential tumour suppressor in NPC, functioning through inhibition of the oncogenic protein metadherin (MTDH). Interestingly, NPC cases with MTDH overexpression exhibited an increased risk of disease recurrence (41). Low miR-451 expression was associated with decreased survival in NPC patients, and has been shown to function by increasing cell growth and invasion by targeting macrophage migration inhibitory factor (MIF) in NPC cells (42). miR-26a, miR-98, and miR-101 have all been reported to function similarly as tumour suppressors in NPC (45). Under-expression of these miRNAs in NPC leads to de-repression of EZH2, causing loss of repression of targets of EZH2 regulation, including c-Myc, cyclins D3 and E2, and cyclin-dependent kinase 4 (CDK4) and CDK6 (43,44). Lastly, miR-216b has been shown to promote NPC growth and invasion by targeting K-RAS (46); conversely, miR-34c has been shown to suppress growth and metastasis by targeting the proto-oncogene MET (47), both of which are significantly downregulated in primary NPC tissues.
Oncogenic microRNAs
Over-expression of a number of oncogenic miRNAs has been described in NPC. miR-18a and miR-18b are members of the miR-17~92 oncogenic miRNA cluster, which has well documented oncogenic functions in various tumour types (76). In NPC, miR-18a is highly over-expressed (70), resulting in direct inhibition of the miRNA biogenesis regulatory protein Dicer1, causing a global downregulation of miRNA expression in NPC (48). miR-18b is also over-expressed in NPC, which has been associated with disease progression and poor outcome. miR-18b functions to repress connective tissue growth factor (CTGF), thereby enhancing cellular proliferation (49).
Over-expression of miR-141 in NPC has been linked with increased cell growth, migration and invasion, as well as loss of cell cycle regulation and reduced apoptosis. This regulation is thought to occur through down-regulation of the putative target genes phosphatase and tensin homolog (PTEN), ubiquitin-associated protein 1 (UBAP1), and bromodomain containing 3 (BRD3) in NPC. Additionally, expression of miR-141 is regulated by the oncogenes c-Myc and short palate, lung, and nasal epithelium clone 1 (SPLUNC1) (50), highlighting the tight regulatory network involving miR-141 in NPC.
Expression of miR-155 stimulates proliferation, migration and invasion by regulation of target genes Jumonji Domain 1A (JMJD1A) and BTB and CNC homology 1 (BACH1), and its expression has been strongly associated with tumour stage and patient survival. Interestingly, regulation of miR-155 occurs through the EBV-encoded LMP1 and LMP2A proteins (51,77). In addition, miR-144 is over-expressed in NPC and also functions to inhibit PTEN expression, causing increased cell proliferation, invasion, and metastasis (52). miR-214 has also been closely linked with increased metastasis in NPC, both in cell lines and primary human samples, functioning at least in part via inhibition of the tumour suppressor lactotransferrin (LTF) (53). Furthermore, miR-214 has been shown to enhance proliferation and promote an anti-apoptotic phenotype in NPC cells (78). In addition to miR-155, miR-144 and miR-214, a number of other miRNAs over-expressed in NPC also contributed significantly to enhancing the metastatic phenotype of NPC. miR-30a has been shown both in vitro and in vivo to increase metastasis and invasion by inhibiting E-cadherin activity (54). Furthermore, expression of miR-149 was elevated in highly metastatic NPC cells, contributing to increased migration, invasion and epithelial-mesenchymal phenotypes through E-cadherin inhibition (55). miR-93 has been reported to inhibit transforming growth factor-β receptor II (TGFβRII) (56) and disabled homolog-2 (DAB2) (57), thereby regulating tumour cell growth invasion and metastasis. Finally, miR-504 was also recently implicated as an oncogenic miRNA in NPC, functioning to directly target nuclear respiratory factor 1 (NRF1), wherein increased expression correlated with poor response to radiation therapy (58).
Epstein-Barr virus (EBV)-associated small RNAs and microRNAs in nasopharyngeal carcinoma (NPC)
The EBV encodes a number of small non-coding RNAs with oncogenic properties, including EBV-encoded small RNAs (EBER) 1 and 2, and BamH1-A region rightward transcript (BART) miRNAs (79). The involvement of these miRNAs and small non-coding RNAs in NPC pathogenesis are detailed in the following section.
Epstein-Barr virus (EBV)-encoded microRNAs
Since the discovery of the first EBV-associated miRNA in 2004 (80), a total of 48 mature miRNAs encoded by EBV in 25 precursor miRNAs have been identified, located in two major coding regions of the virus; BART (encoding 44 mature miRNAs) and the open reading frame of the BHRF1 gene (encoding 4 mature miRNAs) (81,82). Numerous studies have identified over-expression of BART miRNAs in NPC (81,83,84). Interestingly, unlike the BART encoded miRNAs, BHRF1-miRNAs have not been found in EBV-associated NPC tissues (85). EBV-associated miRNAs are known to modulate multiple viral and human mRNAs. For example, BART miRNAs have putative binding sites in numerous EBV mRNA targets, including LMP1 (targeted by miR-BART1, 9, 16, and 17), and LMP2A (targeted by miR-BART22). BART-miRNAs have been functionally shown to modulate cellular proliferation, survival, and evasion of host immunity (86,87).
Human targets of the BART-miRNAs include numerous cancer-associated proteins, such as PTEN (59), p53 up-regulated modulator of apoptosis (PUMA) (60), Bcl-2-interacting mediator of cell death (BIM) (61), determination of interleukin 4 commitment 1 (DICE1) (62), E-cadherin (63), and translocase of outer mitochondrial membrane 22 homolog (TOMM22) (64). In addition, BART miRNAs target a number of genes associated with host immune regulation, including importin 7 (IPO7) (64), dicer (65), and major histocompatibility complex class I-related chain B (MICB) (66).
A recent study reported that miR-BART1 was highly expressed in NPC, and was closely linked with advanced pathological and clinical stage (59). Furthermore, increased miR-BART1 expression resulted in enhanced migration and invasion in NPC cells in vitro and induced tumour metastasis in vivo. Significantly, this study defined the PTEN tumour suppressor as a direct target of miR-BART1. Functionally, miR-BART1 expression was found to drive epithelial–mesenchymal transition as a consequence of PTEN suppression, and subsequent activation of the PI3K-AKT, FAK-p130Cas and Shc-MAPK/ERK1/2 signaling cascades, leading to increased NPC cell migration, invasion and metastasis (59). Additionally, the level of miR-BART17 quantified in the plasma of NPC patients was significantly elevated in comparison to healthy control individuals (67), further indicating the potential of BART miRNAs as biomarkers for NPC. Another recent study identified high extracellular levels of two EBV-BART-miRNAs (miR-BART7 and 13) in the plasma of NPC patients, and although these levels were highly variable between patients, expression was markedly absent in the plasma of both non-NPC and healthy patient controls (68). Importantly, higher levels of miR-BART7 were associated with a more advanced disease stage. Furthermore, plasma levels of both miR-BART7 and 13 were significantly diminished following treatment with radiotherapy, and when used in combination, these miRNAs provided a 90% predictive confidence of NPC outcome. The significant variation in miR-BART7 and 13 between NPC plasma samples however, might well limit their role as predictive biomarkers. Nonetheless, this study demonstrated the potential use of these miRNAs to serve as additional biomarkers for NPC diagnosis and prediction of treatment response, with significant promise for monitoring disease remission and recurrence. Importantly, expression of miR-BART7 was significantly elevated in NPC cells that were resistant to chemotherapeutic treatment with cisplatin (69). Future studies examining the biological functions of miR-BART7 and 13 will be essential to determine the functional relevance of these putative biomarkers. Taken together, EBV-encoded miRNAs play a fundamental role in controlling key disease processes, including modulation of cellular proliferation and survival and regulation of the host immune response in EBV-associated NPC.
EBV-encoded small RNAs (EBERs) (small non-coding RNAs)
In addition to the miRNAs encoded by EBV, the NPC-associated virus also encodes small non-coding RNAs. The EBV-encoded small RNAs EBER1 and EBER2 are clearly implicated in NPC pathogenesis, with evidence supporting a role for EBER1 and EBER2 in enhancing cell growth and survival, and modulating innate immunity in patients (88). Additionally, these small RNAs (167 and 172 nucleotides in length, respectively) are the most abundantly expressed EBV viral transcripts and are expressed at significantly elevated levels in NPC cells, with up to 1 million copies per cell. The proliferation inducing effects of EBER1 and EBER2 are mediated, at least in part, through promotion of insulin-like growth factor-1 (IGF-1) expression through induction of toll-like receptor 3 (TLR3) signaling (88). The link between EBER and IGF-1 is further evidenced by the high level of IGF-1 expression detected in primary NPC biopsies and the dependency of EBV-positive NPC cell line C666-1 cell growth on IGF-1 signaling (88). Recently, EBER was causally linked to NPC development via induction of retinoic acid-inducible gene 1 (RIG-1), a cytosolic protein that detects double-stranded RNA. Upon detection of EBER by RIG-1, a pro-inflammatory response is initiated in NPC cells through activation of canonical transcription factors, including NF-κB and interferon regulator factor 3 (IRF3), which resulted in increased growth for both in vitro and in vivo models of NPC (89). Importantly, this was the first report to link EBER and RIG-1 signaling in NPC.
MicroRNAs as diagnostic and prognostic biomarkers in nasopharyngeal carcinoma
Due to the difficulty in stratifying NPCs into clinically relevant treatment classifications, reliable molecular biomarkers are needed for accurate prognostication. Profiling studies in a variety of cancer types have demonstrated that miRNA expression varies significantly between normal and tumour tissue (90-92). Additionally, miRNA expression signatures are capable of discriminating between tumour sub-types, offering a tool for accurate diagnosis and for differentiating tumours into clinically informative categories. In particular, EBV-encoded miRNAs have been identified as useful biomarkers for NPC diagnosis and screening (68,84). miRNAs also offer significant potential as sensitive and specific biomarkers for cancer prognosis and treatment prediction. Interestingly, several of the miRNAs identified in predictive signatures for NPC and other cancers have been directly associated with enhancing tumour initiation or progression, suggesting that many of these predictive miRNAs are also responsible for driving tumorigenesis. For instance, Liu et al. identified a 5-miRNA prognostic signature (miR-26a, miR-29c, miR-30e, miR-93, and miR-142) for NPC that was significantly associated with overall, disease-free, and distant metastasis-free survival (93). Additionally, our group recently identified a highly accurate, non-overlapping 4-miRNA prognostic signature, comprised of miR-34c, miR-140, miR-154, and miR-449b, also associated with distant metastasis in NPC (94). Furthermore, pathway enrichment analysis of these 4 miRNAs indicated a role in cell cycle regulation, highlighting a potentially important role for markers of cell cycle activation as prognostic indicators in NPC (94). The causal link between miRNAs and tumour initiation and progression further underscores their potential utility as accurate and reliable biomarkers. Additionally, several of the miRNAs identified in these prognostic signatures have also been implicated in NPC pathogenesis, including miR-26a, miR-29c, and miR-34c (as summarized in Table 1), providing additional evidence for their oncogenic importance. Moreover, the significance of miRNAs as predictive biomarkers is further indicated by evidence associating miRNA with chemoresistance and radioresistance in NPC. Specifically, miR-29c (33), miR-504 (58), and the EBV-encoded BART miRNAs (68,69) have been associated with resistance of NPC cells to chemotherapy and radiotherapy, highlighting the potential utility of miRNA signatures as predictive markers. However, though numerous reports have demonstrated prognosticating abilities of miRNA signatures in cancer [reviewed in (95)], miRNA signatures have yet to be employed as biomarkers in the clinical setting. Large-scale validation studies will be essential in driving the use of these miRNA signatures into clinical practice for NPC and other malignancies.
Conclusions
In summary, our knowledge of the role of miRNAs in NPC pathogenesis is continually unfolding. Further insight into the biological effects of miRNAs in NPC pathogenesis will inform our understanding of this complex malignancy by aiding in the identification of essential disease processes to provide useful prognostic biomarkers, and reveal novel therapeutic targets for NPC treatment in the future. Translation of these findings into clinical application will undoubtedly improve disease management for NPC.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Li G, Qiu Y, Su Z, et al. Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing. PLoS One 2013;8:e84486. [Crossref] [PubMed]
- Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet 2014;46:866-71. [Crossref] [PubMed]
- Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol 2015;12:11-26. [Crossref] [PubMed]
- Bertoli G, Cava C, Castiglioni I. Castiglioni, MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015;5:1122-43. [Crossref] [PubMed]
- He Y, Lin J, Kong D, et al. Current State of Circulating MicroRNAs as Cancer Biomarkers. Clin Chem 2015;61:1138-55. [Crossref] [PubMed]
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol 2012;22:79-86. [Crossref] [PubMed]
- Rickinson A. Epstein-Barr virus. Virus Res 2002;82:109-13. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Xue WQ, Qin HD, Ruan HL, et al. Quantitative association of tobacco smoking with the risk of nasopharyngeal carcinoma: a comprehensive meta-analysis of studies conducted between 1979 and 2011. Am J Epidemiol 2013;178:325-38. [Crossref] [PubMed]
- Chang ET, Adami HO. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2006;15:1765-77. [Crossref] [PubMed]
- Stenmark MH, McHugh JB, Schipper M, et al. Nonendemic HPV-positive nasopharyngeal carcinoma: association with poor prognosis. Int J Radiat Oncol Biol Phys 2014;88:580-8. [Crossref] [PubMed]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol 2008;20:214-21. [Crossref] [PubMed]
- Singh SK, Pal Bhadra M, Girschick HJ, et al. MicroRNAs--micro in size but macro in function. FEBS J 2008;275:4929-44. [Crossref] [PubMed]
- Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 2015;15:321-33. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol 2006;13:1097-101. [Crossref] [PubMed]
- Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol 2010;42:1316-29. [Crossref] [PubMed]
- Denli AM, Tops BB, Plasterk RH, et al. Processing of primary microRNAs by the Microprocessor complex. Nature 2004;432:231-5. [Crossref] [PubMed]
- Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509-24. [Crossref] [PubMed]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005;6:376-85. [Crossref] [PubMed]
- Calin GA, Croce CM. Croce, MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 2002;99:15524-9. [Crossref] [PubMed]
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 2007;6:60. [Crossref] [PubMed]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 2006;6:259-69. [Crossref] [PubMed]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006;94:776-80. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- O'Kelly F, Marignol L, Meunier A, et al. MicroRNAs as putative mediators of treatment response in prostate cancer. Nat Rev Urol 2012;9:397-407. [Crossref] [PubMed]
- Westphal M, Lamszus K. Circulating biomarkers for gliomas. Nat Rev Neurol 2015;11:556-66. [Crossref] [PubMed]
- Agulnik M, Epstein JB. Nasopharyngeal carcinoma: current management, future directions and dental implications. Oral Oncol 2008;44:617-27. [Crossref] [PubMed]
- Sam CK, Brooks LA, Niedobitek G, et al. Analysis of Epstein-Barr virus infection in nasopharyngeal biopsies from a group at high risk of nasopharyngeal carcinoma. Int J Cancer 1993;53:957-62. [Crossref] [PubMed]
- Raab-Traub N. Novel mechanisms of EBV-induced oncogenesis. Curr Opin Virol 2012;2:453-8. [Crossref] [PubMed]
- Liu N, Tang LL, Sun Y, et al. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett 2013;329:181-8. [Crossref] [PubMed]
- Zhang JX, Qian D, Wang FW, et al. MicroRNA-29c enhances the sensitivities of human nasopharyngeal carcinoma to cisplatin-based chemotherapy and radiotherapy. Cancer Lett 2013;329:91-8. [Crossref] [PubMed]
- Sengupta S, den Boon JA, Chen IH, et al. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A 2008;105:5874-8. [Crossref] [PubMed]
- Lu J, Luo H, Liu X, et al. miR-9 targets CXCR4 and functions as a potential tumor suppressor in nasopharyngeal carcinoma. Carcinogenesis 2014;35:554-63. [Crossref] [PubMed]
- Gao F, Zhao ZL, Zhao WT, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun 2013;431:610-6. [Crossref] [PubMed]
- Wong TS, Man OY, Tsang CM, et al. MicroRNA let-7 suppresses nasopharyngeal carcinoma cells proliferation through downregulating c-Myc expression. J Cancer Res Clin Oncol 2011;137:415-22. [Crossref] [PubMed]
- Liu Z, Wu K, Yang Z, et al. High-mobility group A2 overexpression is an unfavorable prognostic biomarker for nasopharyngeal carcinoma patients. Mol Cell Biochem 2015;409:155-62. [Crossref] [PubMed]
- Cai K, Wan Y, Sun G, et al. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol Rep 2012;28:2101-6. [PubMed]
- Xia H, Ng SS, Jiang S, et al. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun 2010;391:535-41. [Crossref] [PubMed]
- Hui AB, Bruce JP, Alajez NM, et al. Significance of dysregulated metadherin and microRNA-375 in head and neck cancer. Clin Cancer Res 2011;17:7539-50. [Crossref] [PubMed]
- Liu N, Jiang N, Guo R, et al. MiR-451 inhibits cell growth and invasion by targeting MIF and is associated with survival in nasopharyngeal carcinoma. Mol Cancer 2013;12:123. [Crossref] [PubMed]
- Lu J, He ML, Wang L, et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res 2011;71:225-33. [Crossref] [PubMed]
- Yu L, Lu J, Zhang B, et al. miR-26a inhibits invasion and metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett 2013;5:1223-8. [PubMed]
- Alajez NM, Shi W, Hui AB, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis 2010;1:e85. [Crossref] [PubMed]
- Deng M, Tang H, Zhou Y, et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J Cell Sci 2011;124:2997-3005. [Crossref] [PubMed]
- LLi YQ, Ren XY, He QM, et al. MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis 2015;6: e1618.
- Luo Z, Dai Y, Zhang L, et al. miR-18a promotes malignant progression by impairing microRNA biogenesis in nasopharyngeal carcinoma. Carcinogenesis 2013;34:415-25. [Crossref] [PubMed]
- Yu X, Zhen Y, Yang H, et al. Loss of connective tissue growth factor as an unfavorable prognosis factor activates miR-18b by PI3K/AKT/C-Jun and C-Myc and promotes cell growth in nasopharyngeal carcinoma. Cell Death Dis 2013;4:e634. [Crossref] [PubMed]
- Zhang L, Deng T, Li X, et al. microRNA-141 is involved in a nasopharyngeal carcinoma-related genes network. Carcinogenesis 2010;31:559-66. [Crossref] [PubMed]
- Du ZM, Hu LF, Wang HY, et al. Upregulation of MiR-155 in nasopharyngeal carcinoma is partly driven by LMP1 and LMP2A and downregulates a negative prognostic marker JMJD1A. PLoS One 2011;6:e19137. [Crossref] [PubMed]
- Franceschini N, Fox E, Zhang Z, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet 2013;93:545-54. [Crossref] [PubMed]
- Deng M, Ye Q, Qin Z, et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol 2013;34:1793-800. [Crossref] [PubMed]
- Wang HY, Li YY, Fu S, et al. MicroRNA-30a promotes invasiveness and metastasis in vitro and in vivo through epithelial-mesenchymal transition and results in poor survival of nasopharyngeal carcinoma patients. Exp Biol Med (Maywood) 2014;239:891-8. [Crossref] [PubMed]
- Luo Z, Zhang L, Li Z, et al. miR-149 promotes epithelial-mesenchymal transition and invasion in nasopharyngeal carcinoma cells. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2011;36:604-9. [PubMed]
- Lyu X, Fang W, Cai L, et al. TGFbetaR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer 2014;13:51. [Crossref] [PubMed]
- Xu YF, Mao YP, Li YQ, et al. MicroRNA-93 promotes cell growth and invasion in nasopharyngeal carcinoma by targeting disabled homolog-2. Cancer Lett 2015;363:146-55. [Crossref] [PubMed]
- Zhao L, Tang M, Hu Z, et al. miR-504 mediated down-regulation of nuclear respiratory factor 1 leads to radio-resistance in nasopharyngeal carcinoma. Oncotarget 2015;6:15995-6018. [Crossref] [PubMed]
- Cai L, Ye Y, Jiang Q, et al. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat Commun 2015;6:7353. [Crossref] [PubMed]
- Choy EY, Siu KL, Kok KH, et al. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med 2008;205:2551-60. [Crossref] [PubMed]
- Marquitz AR, Mathur A, Nam CS, et al. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology 2011;412:392-400. [Crossref] [PubMed]
- Lei T, Yuen KS, Xu R, et al. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer 2013;133:79-87. [Crossref] [PubMed]
- Hsu CY, Yi YH, Chang KP, et al. The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog 2014;10:e1003974. [Crossref] [PubMed]
- Dölken L, Malterer G, Erhard F, et al. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe 2010;7:324-34. [Crossref] [PubMed]
- Iizasa H, Wulff BE, Alla NR, et al. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem 2010;285:33358-70. [Crossref] [PubMed]
- Nachmani D, Stern-Ginossar N, Sarid R, et al. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 2009;5:376-85. [Crossref] [PubMed]
- Gourzones C, Ferrand FR, Amiel C, et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients--evidence of non-exosomal transport. Virol J 2013;10:119. [Crossref] [PubMed]
- Zhang G, Zong J, Lin S, et al. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int J Cancer 2015;136:E301-12. [Crossref] [PubMed]
- Chan JY, Gao W, Ho WK, et al. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res 2012;32:3201-10. [PubMed]
- Yuva-Aydemir Y, Simkin A, Gascon E, et al. MicroRNA-9: functional evolution of a conserved small regulatory RNA. RNA Biol 2011;8:557-64. [Crossref] [PubMed]
- Ma L, Young J, Prabhala H, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 2010;12:247-56. [PubMed]
- Gwak JM, Kim HJ, Kim EJ, et al. MicroRNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat 2014;147:39-49. [Crossref] [PubMed]
- Yu T, Liu K, Wu Y, et al. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/beta-catenin signaling pathway. Oncogene 2014;33:5017-27. [Crossref] [PubMed]
- Lu J, Xu X, Liu X, et al. Predictive value of miR-9 as a potential biomarker for nasopharyngeal carcinoma metastasis. Br J Cancer 2014;110:392-8. [Crossref] [PubMed]
- Li T, Chen JX, Fu XP, et al. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep 2011;25:1353-63. [PubMed]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008;133:217-22. [Crossref] [PubMed]
- Zhu X, Wang Y, Sun Y, et al. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Otorhinolaryngol 2014;271:1939-45. [Crossref] [PubMed]
- Zhang ZC, Li YY, Wang HY, et al. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS One 2014;9:e86149. [Crossref] [PubMed]
- Frappier L. Role of EBNA1 in NPC tumourigenesis. Semin Cancer Biol 2012;22:154-61. [Crossref] [PubMed]
- Pfeffer S, Zavolan M, Grässer FA, et al. Identification of virus-encoded microRNAs. Science 2004;304:734-6. [Crossref] [PubMed]
- Chen SJ, Chen GH, Chen YH, et al. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS One 2010;5:e12745. [Crossref] [PubMed]
- Kuzembayeva M, Hayes M, Sugden B. Multiple functions are mediated by the miRNAs of Epstein-Barr virus. Curr Opin Virol 2014;7:61-5. [Crossref] [PubMed]
- Zhu JY, Pfuhl T, Motsch N, et al. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J Virol 2009;83:3333-41. [Crossref] [PubMed]
- Wong AM, Kong KL, Tsang JW, et al. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer 2012;118:698-710. [Crossref] [PubMed]
- Cosmopoulos K, Pegtel M, Hawkins J, et al. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol 2009;83:2357-67. [Crossref] [PubMed]
- Lo AK, To KF, Lo KW, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A 2007;104:16164-9. [Crossref] [PubMed]
- Lung RW, Tong JH, Sung YM, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia 2009;11:1174-84. [Crossref] [PubMed]
- Takada K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Semin Cancer Biol 2012;22:162-5. [Crossref] [PubMed]
- Duan Y, Li Z, Cheng S, et al. Nasopharyngeal carcinoma progression is mediated by EBER-triggered inflammation via the RIG-I pathway. Cancer Lett 2015;361:67-74. [Crossref] [PubMed]
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [Crossref] [PubMed]
- Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol 2008;26:462-9. [Crossref] [PubMed]
- Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257-61. [Crossref] [PubMed]
- Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol 2012;13:633-41. [Crossref] [PubMed]
- Bruce JP, Hui AB, Shi W, et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget 2015;6:4537-50. [Crossref] [PubMed]
- Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst 2012;104:528-40. [Crossref] [PubMed]


