Editor’s note:
The special column “Statistics in Oncology Clinical Trials” is dedicated to providing state-of-the-art review or perspectives of statistical issues in oncology clinical trials. Our Chairs for the column are Dr. Daniel Sargent and Dr. Qian Shi, Division of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, MN, USA. The column is expected to convey statistical knowledge which is essential to trial design, conduct, and monitoring for a wide range of researchers in the oncology area. Through illustrations of the basic concepts, discussions of current debates and concerns in the literature, and highlights of evolutionary new developments, we are hoping to engage and strengthen the collaboration between statisticians and oncologists for conducting innovative clinical trials. Please follow the column and enjoy.
Statistical aspect of translational and correlative studies in clinical trials
Introduction
Biomarkers play a prominent role in cancer research and development. Gene expression microarrays and single nucleotide polymorphism arrays were commonly used technologies in earlier research. Today, a platform such as next generation sequencing is often used. This tool can be used to measure gene expression, RNA-Seq, methylation, TF binding Chip-Seq, and genetic variant discovery and quantification. Most of these data are generated from Illumina (Solexa), 454 Roche, and SOLiD sequencing machines. In patients care, biomarkers can potentially be used for risk stratification in terms of clinical outcome and may assist physicians in making treatment decisions. Apart from genetic biomarkers, imaging biomarkers can also serve as a potential surrogate for clinical trial endpoints, or guide the treatment routine. These biomarkers are being integrated into many modern clinical trials (1).
In the discovery and identification of biomarkers from big ‘-omics’ data for clinical outcomes, application of sound statistical approaches is essential. We will discuss several statistical issues and introduce statistical methods and strategies for consideration. Without proper implementation of these steps, the resources spent on designing and running an independent clinical validation may turn out to be unfruitful. In this article, we will define some of the terminologies commonly used, discuss how to build and evaluate classifiers, and describe strategies to validate them retrospectively and prospectively.
Definitions
Biomarkers can mainly be classified into three different groups, depending on their intended use in treatment. The evaluation requirements and validation criteria vary according to the purpose of the usage of the biomarkers.
- Prognostic biomarkers, which are associated with patients’ overall outcome. A validated prognostic biomarker provides the opportunity to identify patients at high risk and thus a population that may benefit from early or aggressive intervention. For example, KRAS mutation is associated with poor prognosis in non-small cell lung cancer (NSCLC) patients (2).
- Predictive biomarkers, which predict the effect of a specific treatment on a clinical endpoint for patients. As an example, advanced pancreatic cancer patients with lower levels of vascular endothelial growth factor-D (VEGF-D) benefited from the addition of bevacizumab to standard gemcitabine, while patients with high VEGF-D levels did not (3). Another example is that patients with overexpressed Cyclo-oxygenase-2 (COX-2) who appeared to benefit from the addition of celecoxib (a COX-2 inhibitor) to standard chemotherapy relative to those receiving chemotherapy only (4).
- Biomarkers which can potentially serve as a surrogate for the primary endpoint in clinical trials. Analogous to surrogate clinical endpoints (5), surrogate biomarkers can be used as intermediate indicators of treatment efficacy in cancer treatment studies. For example, maximal pain intensity, an individual measure, on the Brief Pain Inventory quality-of-life instruments in the previous 24 hours, has been used as a surrogate endpoint for clinical benefit (6).
Development of cancer biomarkers: planning and design
Clustering or cluster analysis is an algorithm that can be applied to identify groupings of genes or patients. While it is an excellent discovery tool for unsupervised learning, heatmap and clustering methods applied to a genomic feature set do not rigorously define a classifier, defined as a tool that utilizes a patient’s genetic characteristics to determine which class or group he/she belongs to. Many traditional statistical methods are not capable of handling the large number of genes and small sample size problems that biomarker discovery often encounters. Therefore, modified and new methods are needed for tackling big ‘-omics’ data problems.
Building a classifier
To build a classifier of a clinical outcome based on the pattern of thousands of biomarkers, such as genes or genetic variants, one often uses supervised learning methods to train the classifier with a data set in which true phenotypes of the outcome is known. Classifiers using different supervised learning algorithms have been proposed, including discriminant analysis, decision trees, random forests, nearest neighbor classifiers, neural networks, and support vector machine classifiers. However, there is no consensus in the statistical and machine learning communities about which particular classifier is superior to others across different data sets. Key considerations for deciding which approach might be more appropriate include the ability to handle missing and/or noisy data, interpretability, and predictive power.
Feature selection
Feature selection is a critical component of building a classifier. In our context, genes are the features that require selection. A good classifier depends on the selection of important features, i.e., features that can help distinguish between the categorical outcomes of interest. As in model building, good classifiers that are parsimonious are easier to interpret. Complicated classifiers with too many features can degrade the performance of the classifier and make external validation more difficult. One can utilize univariate test statistics like the two-sample t-test or Wilcoxon rank sum test for all features based on the training set, and then identify the top features by ranking the P values. A classifier is then built on these top features based on the training set. In the survival setting, one may use Cox regression or non-parametric methods to identify top features. Considering the complex relationship of biomarkers with the associated phenotype, one often believes a decision based on multiple biomarkers may potentially be more useful than individual biomarkers. There have also been methods developed to identify multiple genes such as the approach developed by Pang et al. [2012] for survival outcomes (7).
Strategies for internal validation
Overfitting happens when the model corresponds too closely to a particular data set. As a result, the model may not predict future observations well. To prevent overfitting the data, validation methods such as cross-validation can be employed. Internal validation uses the data set from the same set of patients as was used to develop the classifier to assess the performance of the classifier. To ensure an unbiased evaluation, one must ensure that the data used for evaluating the predictive accuracy of the classifier be distinct from the data used for selecting the biomarkers and building the supervised classifier. This can be achieved by resampling techniques including hold-out or split sample, k-fold cross validation and leave-one-out cross validation. The hold-out method is usually applied to larger data sets, while the leave-one-out cross validation may provide the best option for smaller data sets. K-fold cross validation with k=5 or 10 are commonly used for various sizes of data. Some investigators will also incorporate permutation and nested cross validation strategies. Other strategies to help reduce overfitting include dimension reduction, penalization, and the use of Bayesian methodology.
Retrospective validation
After the classifier is built, the next step is to perform retrospective validation, i.e., validation based on existing clinical data and samples. These samples are independent from the original training data in the previous step. A locked down model should be pre-specified. This model is then used to predict the outcome of interest in the independent validation data. The predicted outcome is then compared against true clinical outcomes for concordance and/or accuracy. However, this may not always be possible. Large databases of ‘-omics’ data may turn out to be too heterogeneous for validation, or the patient population may turn out to be different from that used in model-building. Moreover, investigators may face issues such as assay platform changes or differences in sample collection protocol. Despite these potential drawbacks, some researchers turn to biospecimen banks, where samples have been collected from large clinical trials (8), such as the NCI National Clinical Trials Network. One such example is the CALGB 140202 lung cancer tissue bank (9) that has contributed samples to multiple studies, including microRNA signature validation, gene-expression signature validation, The Cancer Genome Atlas (http://cancergenome.nih.gov/), exome-sequencing, blood biomarkers, and protein assay validation.
Sample size calculation
Researchers have taken different strategies in sample size calculations for designing studies assessing ‘-omics’ data. Jung [2005] described an approach for sample size calculation based on false discovery rate control in microarray data analysis (10). Dobbin and Simon [2007] provided a sample size calculation algorithm based on the specification of some level of tolerance within its true accuracy (11). Pang and Jung [2013] developed a sample size calculation method that may be used to design a validation study from pilot data (12). These sample size calculation methods require the knowledge of the expected effect sizes, number of genes on the platform, sample proportions, the desired level of statistical power, and the acceptable type I error or false discovery rate.
Pathway analysis
Biology is generally not dictated by a single gene, but rather a set of genes. Pathways are set of genes that serve different cellular or physiologic functions. Pathways are becoming more important in identifying biomarkers and molecular targets for diagnosis and treatment. These pathways can come from pathway databases such as KEGG or Gene Ontology. In recent years, researchers have developed methods to associate gene expression or single nucleotide polymorphisms with prognosis and identify gene signatures (13,14). Statistical methods for pathway analysis based on machine learning, Bayesian approaches and enrichment tests have been developed in the past few years. These pathway-based approaches allow scientists to focus on limited sets of genes, select targets from multiple biomarkers, and gain insights into the biological mechanisms of the tumor. Using random forests importance measure, one can select features in a pathway-based setting (13,15). Compared to single -gene based analysis, pathway-based methods can identify more subtle changes in expression (16).
Evaluation strategies
To evaluate the accuracy of predicting a binary outcome based on a classifier with two statuses, we often consider the use of a 2 by 2 table. This table is often called the confusion table. The sum of the diagonal values divided by the total number of participants indicates the prediction or classification accuracy. Several other measures based on true positive (TP), true negative (TN), false positive (FP), and false negative (FN), are also important for consideration. Using the values in cells labelled as A, B, C, D in Table 1, these measures can be defined as: (I) positive predictive value (PPV) = A/(A + C); (II) negative predictive value (NPV) = D/(B + D); (III) sensitivity = A/(A + B); and (IV) specificity = D/(C + D). The Area Under the receiver operating characteristics (ROC) Curve (AUC) is also commonly used. A value of 0.5 represents a random guess while a 1 represents a perfect prediction.
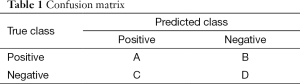
Full table
One approach to assess survival prediction performance is to compare the predicted survival of various risk groups using a log-rank test. This can be coupled with permutation testing when appropriate. To evaluate the accuracy of survival prediction without dichotomizing, we can employ the area under the ROC curve (AUC) approach for survival data of Heagerty et al. [2000] (17). In this instance, sensitivity and specificity are defined as a function of time, and the time-dependent ROC curve is a plot of sensitivity (t) versus 1—specificity (t). Higher prediction accuracy is supported by a larger AUC value. An alternative would be to use the concordance index (C-index) (18), a measure of how well the prediction algorithm ranks the survival of any pair of individuals. C-index takes values between 0 and 1. A C-index of 0.5 corresponds to a random guess and 1 means perfect concordance.
Prospective trial designs
We briefly discuss three main types of designs for prospective validation: targeted design, biomarker-stratified randomized (BSR) design, and hybrid design. Additional details can be found in Simon 2014 (CCO) (19).
Targeted design
For a targeted design, a biomarker is used to restrict eligibility for a randomized clinical trial comparing an experimental regimen to standard of care or control. Often, the experimental regimen is a targeted agent developed for those patients with a particular mutational status of a biomarker. When evaluating the treatment efficacy of a target agent using a randomized phase III trial, the targeted design can be much more efficient than untargeted design. However, a targeted design prevents the chance to test for interaction between treatment and the biomarker. It also prevents the researcher from validating the performance of the predictive biomarker by restricting enrollment to marker-positive only patients. CALGB 30801 is a good example of such design to validate the findings from CALGB 30203 in which patients whose tumors over-expressed COX-2 were randomized to either celecoxib or placebo (20).
BSR design
In BSR designs, biomarker status is a stratification factor. For example, both marker positive and negative patients are randomized to a targeted agent versus standard of care or placebo, with randomization stratified by biomarker status. The BSR design allows testing of whether the marker positive patients benefit from an agent compared to standard of care or placebo, with randomization stratified by biomarker status. testing of an overall treatment benefit, and an evaluation of the predictive classifier’s performance in identifying the targeted subgroup of patients. However, the drawback of BSR design is the resources and time needed for the conduct of the trial. The ability to answer several questions comes at a cost of the need of more treated patients, and potentially longer follow-up. If the overall treatment benefit is small and the patient population is predominately marker negative, such a design can be ineffective and unethical for the marker negative patients. However, the BSD does avoid a limitation of the following design (hybrid design) that one must be highly confident that the biomarker can identify the subgroup of patients who may benefit.
Hybrid design
A hybrid design lies between targeted and BSR designs. Like the BSR design, the hybrid design randomizes both marker-positive and marker-negative patients. But to reduce cost and improve study efficiency, for example, only a subset of all marker-negative patients is randomized. The process of selecting which patients to randomize may depend on biomarker prediction, clinical outcome, or other baseline patients’ characteristics. The efficiency gain due to a hybrid design could be significant when marker negative patients are predominant in the unselected patient population and auxiliary variables exist to identify those informative patients. If the targeted therapy benefits a subgroup of the patient population, but the biomarker used does poorly in the identification of the group, then a useful therapy could be halted for further investigation. An example of the hybrid design is EORTC 10041 (21), which restricted eligibility to only node-negative breast cancer patients to assess a 70-gene expression profile developed by the Netherlands Cancer Institute.
Reporting guidelines
The Strategy Group of the Program for the Assessment of Clinical Cancer Tests and a working group of a National Cancer Institute-European Organization Research Treatment Collaboration developed the Reporting recommendations for tumor Marker prognostic studies (REMARK) (22,23). Many high profile journals require that submissions be vetted through this guideline. This guideline provides a thorough 20-item checklist on essential pieces in the publication of marker-based studies, such as assay methods, study design, and statistical methods. It also focuses on presentation of the study results, with guidelines for data, analysis and presentation.
Discussion
The availability of big ‘-omics’ data presents an exciting opportunity for researchers to translate their findings and discovery into clinical trials and ultimately clinical practice. Presently, biomarker discovery is an integral part of the main clinical study. Special attention in planning the study at the protocol development stage can help facilitate testing of secondary hypotheses, collection of specimens, and statistical analysis. While we have covered multiple aspects of statistical considerations for correlative studies in clinical trials, some important topics not covered include differentially expressed genes (DEGs), prospectively validation study designs for prognostic markers, and multiple hypothesis testing issues. Additionally, specific cancers may have their unique topics (24). As sequencing becomes more affordable, we expect that biomarkers will become a routine component of clinical trials. The big ‘-omics’ data generated from these technologies will prove invaluable in this personalized medicine era.
Acknowledgements
Funding: This work is partially supported by National Institute of Health’s National Cancer Institute under the award P01CA142538.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hall JA, Salgado R, Lively T, et al. A risk-management approach for effective integration of biomarkers in clinical trials: perspectives of an NCI, NCRI, and EORTC working group. Lancet Oncol 2014;15:e184-93. [PubMed]
- Zhu CQ, da Cunha Santos G, Ding K, et al. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 2008;26:4268-75. [PubMed]
- Nixon AB, Pang H, Starr MD, et al. Prognostic and predictive blood-based biomarkers in patients with advanced pancreatic cancer: results from CALGB80303 (Alliance). Clin Cancer Res 2013;19:6957-66. [PubMed]
- Edelman MJ, Watson D, Wang X, et al. Eicosanoid modulation in advanced lung cancer: cyclooxygenase-2 expression is a positive predictive factor for celecoxib + chemotherapy--Cancer and Leukemia Group B Trial 30203. J Clin Oncol 2008;26:848-55. [PubMed]
- Sargent DJ, Wieand HS, Haller DG, et al. Disease-free survival versus overall survival as a primary end point for adjuvant colon cancer studies: individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol 2005;23:8664-70. [PubMed]
- Atkinson TM, Mendoza TR, Sit L, et al. The Brief Pain Inventory and its “pain at its worst in the last 24 hours” item: clinical trial endpoint considerations. Pain Med 2010;11:337-46. [PubMed]
- Pang H, George SL, Hui K, et al. Gene selection using iterative feature elimination random forests for survival outcomes. IEEE/ACM Trans Comput Biol Bioinform 2012;9:1422-31.
- Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446-52. [PubMed]
- Sugarbaker D, Richards W, Wang X, et al. CALGB 140202 The CALGB Lung Cancer Tissue Bank. April 2011. Protocol. Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=649826&version=healthprofessional
- Jung SH. Sample size for FDR-control in microarray data analysis. Bioinformatics 2005;21:3097-104. [PubMed]
- Dobbin KK, Simon RM. Sample size planning for developing classifiers using high-dimensional DNA microarray data. Biostatistics 2007;8:101-17. [PubMed]
- Pang H, Jung SH. Sample size considerations of prediction-validation methods in high-dimensional data for survival outcomes. Genet Epidemiol 2013;37:276-82. [PubMed]
- Pang H, Datta D, Zhao H. Pathway analysis using random forests with bivariate node-split for survival outcomes. Bioinformatics 2010;26:250-8. [PubMed]
- Pang H, Hauser M, Minvielle S. Pathway-based identification of SNPs predictive of survival. Eur J Hum Genet 2011;19:704-9. [PubMed]
- Pang H, Lin A, Holford M, et al. Pathway analysis using random forests classification and regression. Bioinformatics 2006;22:2028-36. [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [PubMed]
- Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337-44. [PubMed]
- Harrell FE Jr, Califf RM, Pryor DB, et al. Evaluating the yield of medical tests. JAMA 1982;247:2543-6. [PubMed]
- Simon R. Biomarker based clinical trial design. Chin Clin Oncol 2014;3:39. [PubMed]
- Edelman MJ, Wang X, Hodgson L, et al. Phase III randomized, placebo controlled trial of COX-2 inhibition in addition to standard chemotherapy for advanced NSCLC: CALGB 30801 (Alliance). American Association for Cancer Research (AACR) annual meeting. San Diego, CA, 2014.
- Viale G, Slaets L, Bogaerts J, et al. High concordance of protein (by IHC), gene (by FISH; HER2 only), and microarray readout (by TargetPrint) of ER, PgR, and HER2: results from the EORTC 10041/BIG 03-04 MINDACT trial. Ann Oncol 2014;25:816-23. [PubMed]
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med 2012;9:e1001216. [PubMed]
- McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor MARKer prognostic studies (REMARK). Nat Clin Pract Urol 2005;2:416-22. [PubMed]
- Glickman M, Wang X, Pang H, et al. Building and Validating High Throughput Lung Cancer Biomarkers. Chance 2009;22:55-62.


