Genotyping of mucosal melanoma
Introduction
Mucosal melanomas are characterized by an aggressive biological behavior. Melanoma has historically been refractory to traditional therapeutic approaches. The high mortality rate of mucosal melanoma has essentially been due to the lack of effective traditional therapeutic approaches. Significant improvements to existed therapies or the development of new drugs are expected to increase survival of patients with mucosal melanoma.
A growing understanding of the biology and molecular mechanisms of melanoma has led to the identification of a number of driver mutations for this aggressive tumor. Recent successes of molecularly based targeted drugs in metastatic melanoma can be attributed to the identification of the genetic alterations in these melanoma targets. This review summarizes the current and emerging molecular targets in mucosal melanoma, and discusses the potential application of these molecules in targeted therapy.
Whole genome sequencing of mucosal melanoma
All melanocytes share the same embryologic origin, but microenvironment shaping their final destinations in different sites of the body differs a lot. Epidermal and dermal melanocytes, as well as melanocytes of the mucosal membranes and uvea, are situated in different kinds of tissues and surrounded by different types of cells. It may be expected that they also differ in signal pathways involved in their growth and maintenance, and consequently differ in the development of melanoma. Accordingly, Aoki et al. suggested the existence of two distinct types of melanocytes in mouse, based on differential signaling requirements for the maintenance of dermal versus epidermal melanocytes (1).
Furney et al. used whole genome and whole exome sequencing to characterize the somatic alterations and mutation spectra in the genomes of ten mucosal melanomas. They showed that mucosal melanomas are characterized by a relatively low mutational burden. The samples analyzed by whole genome sequencing harbored averagely 8,193 somatic single nucleotide variants (SNVs) per mucosal melanoma, while an average of 86,495 SNVs were detected per cutaneous melanoma. Similarly, they observed an average 69 non-synonymous SNVs (nsSNVs) in coding region by whole genome sequencing and 66 nsSNVs by exome sequencing, which was remarkably fewer than the average 375 nsSNVs for sun-exposed melanomas. Thus, the mutational burden in mucosal melanoma is 5- to 10-fold lower than that in cutaneous melanoma. In addition, the cohort showed a much higher rate of copy number and structural variants in mucosal melanoma as compared with cutaneous melanoma (2). Several other studies have also shown that melanomas of mucosal membranes have distinctive patterns of chromosomal aberrations (3-5). These studies provide genetic evidence for the existence of distinct molecular pathways in mucosal melanoma, and indicate the classification of mucosal melanoma as a distinct entity.
RAS/RAF/mitogen activated protein kinase (MAPK) pathway
The RAS-RAF-MEK-ERK MAP kinases pathway plays a central role in the biology of various cell types, and regulates the proliferation of melanocytes (6) (Figure 1). Stimulation of different membrane-bound receptors, mainly receptor tyrosine kinases and G-protein coupled receptors, promotes activation of RAS and then RAF kinases. Activated RAF sequentially activates MEK and ERK, leading to activation of multiple cytoplasmic and nuclear molecules involved in regulating cell proliferation, differentiation and survival (7).
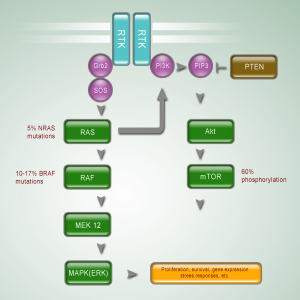
BRAF
The serine-threonine protein kinase RAF has three isoforms: ARAF, BRAF, and CRAF (RAF1). Strikingly over half of the tested melanoma tumors and cell lines had a mutation in the BRAF gene. The high prevalence of BRAF mutations in cutaneous melanoma has subsequently been validated in multiple studies. A recent meta-analysis of sequencing results from over 200 studies, including over 2,700 samples, identified a mutation rate of 65% in melanoma cell lines and 42% in uncultured cutaneous melanomas (8). Mutations in BRAF are the most common somatic mutations in cutaneous melanoma. Over 90% of the identified mutations in BRAF affect the valine residue at position 600, and most of these mutations frequently result in the substitution of a glutamic acid (V600E). However, BRAF mutations are extremely rare in mucosal melanoma. Maldonado et al. find only 10% (2 of 21 patients) BRAF mutations in mucosal membranes (9). The mutation rate of BRAF is even lower in other researches (10,11). Omholt et al. show that the mutation rate in mucosal melanoma is only 6% (12), and Beadling et al. find no BRAF mutation in 45 mucosal melanoma patients (13). One retrospective study in Peking University Cancer Hospital reported the incidence of somatic mutations within BRAF in mucosal subtype is 17.0% (9/53). Among patients with BRAF mutations, 77.8% (7/9) of them bear V600E mutations. Anorectal mucosa is the most common (20.0%) primary sites for patients bearing BRAF mutation. The initial characterization of the prevalence of BRAF mutations is mainly conducted in cutaneous melanomas, but BRAF mutations in mucosal melanomas are relatively rare.
NRAS
RAS family of small GTPases, a family of guanine-nucleotide binding proteins embedded in the inner surface of cell membrane, represents the upstream component of the RAS/RAF/MAPK pathway. Mutations in RAS family members are one of the most common activating events in most of cancers. Members of RAS family mainly include three isoforms: NRAS, KRAS, and HRAS. While mutations in KRAS and HRAS do not appear to be significant in melanoma, mutations in NRAS were identified in melanoma in 1985, even earlier than the discovery of BRAF mutations (14). NRAS mutations lead to retain NRAS in its active GTP-bound state, resulting in aberrant downstream activation of MAPK pathway through RAF (predominantly C-RAF) (15). However, NRAS can also activate other pathways such as the phosphatidylinositol 3-kinases (PI3K) pathway (6). The prevalence of NRAS mutations varies between different clinical-pathologic types of melanoma, although not as dramatically as that observed with BRAF mutations. In Dr. Bastian’s study of the molecular characteristics of melanoma subtypes, NRAS mutation rate was 22% in non-chronic sun-damaged disease (non-CSD) cutaneous melanoma, 15% in CSD cutaneous melanoma, 5% in mucosal melanoma, and 10% in acral melanoma (5). Other studies reported the frequency of NRAS mutations in mucosal subtype was 10-7% (12,16). In 7 NRAS mutations identified by Omholt et al., 4 involved codon 61, and 3 involved codons 12 and 13. Notably, vaginal melanomas showed a NRAS mutation rate of 43%, suggesting that the NRAS mutation rate may be different between sites of mucosa (12). The somatic mutation rate of NRAS gene, reported by Peking University Cancer Hospital, is 5.7% (3/53), and the mutations predominantly affect codon 61.
PI3K-AKT pathway
The PI3K-AKT pathway is one of the most important signaling networks in cancer (Figure 1). Aberrant expression and activity of the components in PI3K-AKT pathway has been shown to promote melanoma genesis (17,18). The PI3K-AKT signaling cascade regulates initiation, progression and invasion of melanoma cells by inhibiting cell senescence and apoptosis, and by inducing cell survival cascades in melanoma cells.
Activation of the PI3K-AKT pathway occurs frequency in cutaneous melanomas, particularly those without CSD (5). Phosphatase and tensin homologue (PTEN), a negative regulator of the pathway, appears to be a common somatic target in melanoma, residing in a commonly deleted genomic area on chromosome 10q. Approximately 20% to 40% of cutaneous melanomas show loss or altered expression of the PTEN tumor suppressor, which acts as an upstream inhibitor of the PI3K-AKT pathway because of its phosphatase activity (19-21). Activating mutations in other components of the PI3K-AKT pathway appear to be quite rare in melanoma. Activating mutations of the catalytic subunit of PI3K and activating mutations of AKT homologous have been detected in ~3% of tested samples (22,23).
The role of PTEN in mucosal melanoma has not been elucidated. However, a recent study reported loss of PTEN expression in 50% of sinonasal melanomas (10). Curtin’s study showed mucosal melanoma had a significantly higher level of loss or altered expression of the PTEN than other subtypes (5). Peking University Cancer Hospital screened activating alterations of PI3K-AKT signaling pathway in 77 mucosal melanoma patients. Immunohistochemistry assays were used to evaluate phosphorylation of mTOR, S6RP, 4E-BP1 and AKT. There are 52 patients (67.5%) showing activation of PI3K-AKT signaling pathway. These researches suggest that activation of PI3K-AKT may be prevalent in mucosal melanoma.
CDKN2A pathway
The cell cycle is propelled by a series of protein kinases: cyclin as the regulatory unit, and cyclin-dependent kinase (CDK) as its catalytic partner. Cyclins, with the bound and activated CDKs, function in distinct stages of the cell cycle. Cyclin/CDK complexes phosphorylate specific protein substrates to regulate the cell cycle. Cyclin/CDK synthesis and subsequent proteolysis ensure the well-defined transitions between stages of the cell cycle (24). The p16-cyclin D/Cdk-pRb-E2F pathway has been implicated in the regulation of cell cycle, as evidenced by frequent aberrations of related components in melanoma (Figure 2). Mutation in CDKN2A is detected in 10-15% of sporadic melanoma patients (25). Hsieh et al. showed that cyclin D1 is recurrently amplified and overexpressed in oral mucosa melanomas, with 61.76% of samples positive for cyclin D1 (26). Curtin et al. found that mucosal melanomas more commonly showed loss of CDKN2A locus and amplifications of CDK4 than melanomas on skin with chronic sun-induced damage or melanomas on skin without chronic sun-induced (5). Thus, the majority of mucosal melanomas does not harbor mutations in BRAF or NRAS, but instead shows increased numbers of copies of downstream gene cyclin D1 or CDK4.
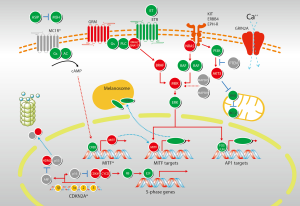
C-KIT
While BRAF and NRAS mutations appear to be common and functionally significant in cutaneous melanomas, their low prevalence in acral, mucosal and uveal melanomas has spurred investigations to identify other genetic events in these subtypes. Beadling et al. screened 189 melanomas for mutations in exons 11, 13, and 17 of C-KIT and detected C-KIT mutations in 23% (3 of 13) of acral melanomas, 15.6% (7 of 45) of mucosal melanomas, 7.7% (1 of 13) of conjunctival melanomas, 1.7% (1 of 58) of cutaneous melanomas, and 0% (0 of 60) of choroidal melanomas (13). Curtin et al., who examined 102 primary melanomas excised from various anatomical sites, found mutations and copy number increases of C-KIT in 39% of mucosal melanomas (11). In Peking University Cancer Hospital’s cohort of 502 Chinese patients, somatic mutations within the C-KIT gene were 10.8% (54 of 502). The frequency of mucosal subtype was 33% (167 of 502). In this cohort, the most frequent C-KIT mutations occurred in exon 11 (27). Rate of C-KIT mutation may be different between arising sites, as Omholt et al. showed that C-KIT mutations were detected in 35% (8 of 23) of vulvar, 9% (2 of 22) of anorectal, 7% (1 of 14) of nasal cavity, and 20% (1 of 5) of penile melanomas (12). Rate of increased C-KIT amplification (up to 31%) was higher than rate of mutations in mucosal melanomas (28).
GNAQ
GNAQ encodes a subunit of the G-protein-coupled receptor. Expression of the GNAQ/GNA11 protein resulted in aberrant activation of MAPK, enhanced anchorage-independent growth, and increased tumorigenicity of melanocytes (Figure 2). More recently, it is found that mutations in exon 5 (Q209) of GNAQ and GNA11 occur frequently in uveal melanomas (29). The GNA11 and GNAQ mutations are mutually exclusive. Furthermore, the Q209 GNA11 mutation was more frequently detected in uveal melanoma metastases (57%) compared with the Q209 GNAQ mutation (22%) (30). Early evidence showed that GNA11 alterations may be more predictive indicator for metastases and thus may represent a potentially crucial therapeutic target. But no GNAQ- or GNA11-targeted therapies currently exist for melanomas harboring GNAQ or GNA11 mutations. Mutation rates of GNAQ and GNA11 are uncertain in mucosal melanoma. Several researches showed that GNA11 mutation, but not GNAQ mutation, may be the major alteration in mucosal melanoma (16). Hopefully an improved understanding of the consequences of these mutations will lead to new and more effective treatments for mucosal melanomas.
MITF
MITF (microphthalmia-associated transcription factor) is the ‘master regulator’ of differentiation, survival and proliferation of normal melanocytes; and is critical in controlling the proliferation, migration and invasion of melanoma cells (31,32) (Figure 2). MITF plays potential role as a melanoma oncogene, as high-level and focal amplification of the MITF locus can be found in approximately 20% of melanomas (33). But the mutation rate in mucosal melanoma subtype has not been defined. Knockdown of MITF with a siRNA induces apoptosis in these MITF-amplified tumors, as well as in many melanomas lacking MITF amplification. It seems that MITF contributes to melanoma pathophysiology even when it is not highly expressed. Many transcription factors and signaling pathways that positively and negatively regulate MITF expression have been elucidated. MITF expression seems to be partially under the control of oncogenic BRAF (34). Specifically, it has been observed that BRAF inhibition results in increased MITF levels, which is consistent with the study showing that MAPK directly phosphorylates MITF and lead to its ubiquitin-dependent proteolysis (35). Understanding the full scope of tumor cell heterogeneity, as well as the related signaling molecules and transcription factors may help the design of secondary therapies that would improve the efficiency of the currently used BRAF inhibitors and circumvent the technical challenge of directly targeting MITF.
Conclusions
In recent years, the classification of melanoma has dramatically changed with the identification of targetable mutations in this disease. While the discovery and characterization of BRAF, NRAS, PTEN, KIT, MITF and GNAQ mutations have definitely improved our understanding of melanoma. Mucosal melanoma shows higher activation of PI3K-AKT signaling pathway and CDKN2A pathway, and harbors higher rate of mutations in c-KIT than that in RAS or BRAF (Table 1). Thus, this subtype of melanomas would be less likely to respond to therapeutic interventions that target upstream components of the MAPK pathway including BRAF. The identification and characterization of genetic aberrations in these patients may help to develop targeted therapy in this subtype. But tremendous effects are required to elucidate the molecular basis of mucosal melanoma.

Full table
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Aoki H, Yamada Y, Hara A, et al. Two distinct types of mouse melanocyte: differential signaling requirement for the maintenance of non-cutaneous and dermal versus epidermal melanocytes. Development 2009;136:2511-21. [PubMed]
- Furney SJ, Turajlic S, Stamp G, et al. Genome sequencing of mucosal melanomas reveals that they are driven by distinct mechanisms from cutaneous melanoma. J Pathol 2013;230:261-9. [PubMed]
- Bastian BC, Olshen AB, LeBoit PE, et al. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol 2003;163:1765-70. [PubMed]
- Van Dijk M, Sprenger S, Rombout P, et al. Distinct chromosomal aberrations in sinonasal mucosal melanoma as detected by comparative genomic hybridization. Genes Chromosomes Cancer 2003;36:151-8. [PubMed]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135-47. [PubMed]
- Sullivan RJ, Flaherty K. MAP kinase signaling and inhibition in melanoma. Oncogene 2013;32:2373-9. [PubMed]
- Wellbrock C, Hurlstone A. BRAF as therapeutic target in melanoma. Biochem Pharmacol 2010;80:561-7. [PubMed]
- Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat 2007;28:578-88. [PubMed]
- Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst 2003;95:1878-90. [PubMed]
- Turri-Zanoni M, Medicina D, Lombardi D, et al. Sinonasal mucosal melanoma: Molecular profile and therapeutic implications from a series of 32 cases. Head Neck 2013;35:1066-77. [PubMed]
- Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340-6. [PubMed]
- Omholt K, Grafström E, Kanter-Lewensohn L, et al. KIT pathway alterations in mucosal melanomas of the vulva and other sites. Clin Cancer Res 2011;17:3933-42. [PubMed]
- Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res 2008;14:6821-8. [PubMed]
- Padua RA, Barrass NC, Currie GA. Activation of N-ras in a human melanoma cell line. Mol Cell Biol 1985;5:582-5. [PubMed]
- Dumaz N, Hayward R, Martin J, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res 2006;66:9483-91. [PubMed]
- Minor DR, Kashani-Sabet M, Garrido M, et al. Sunitinib therapy for melanoma patients with KIT mutations. Clin Cancer Res 2012;18:1457-63. [PubMed]
- Stahl JM, Cheung M, Sharma A, et al. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res 2003;63:2881-90. [PubMed]
- Manning BD, Cantley LC. AKT/PKB AKT/PKB signaling: navigating downstream. Cell 2007;129:1261-74. [PubMed]
- Pollock PM, Walker GJ, Glendening JM, et al. PTEN inactivation is rare in melanoma tumours but occurs frequently in melanoma cell lines. Melanoma Res 2002;12:565-75. [PubMed]
- Tsao H, Zhang X, Benoit E, et al. Identification of PTEN/MMAC1 alterations in uncultured melanomas and melanoma cell lines. Oncogene 1998;16:3397-402. [PubMed]
- Mikhail M, Velazquez E, Shapiro R, et al. PTEN expression in melanoma: relationship with patient survival, Bcl-2 expression, and proliferation. Clin Cancer Res 2005;11:5153-7. [PubMed]
- Curtin JA, Stark MS, Pinkel D, et al. PI3-kinase subunits are infrequent somatic targets in melanoma. J Invest Dermatol 2006;126:1660-3. [PubMed]
- Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer 2008;99:1265-8. [PubMed]
- Israels ED, Israels LG. The cell cycle. Oncologist 2000;5:510-3. [PubMed]
- Hilliard NJ, Krahl D, Sellheyer K. p16 expression differentiates between desmoplastic Spitz nevus and desmoplastic melanoma. J Cutan Pathol 2009;36:753-9. [PubMed]
- Hsieh R, Nico MM, Coutinho-Camillo CM, et al. TThe CDKN2A and MAP kinase pathways: molecular roads to primary oral mucosal melanoma. Am J Dermatopathol 2013;35:167-75. [PubMed]
- Kong Y, Si L, Zhu Y, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res 2011;17:1684-91. [PubMed]
- Yun J, Lee J, Jang J, et al. KIT amplification and gene mutations in acral/mucosal melanoma in Korea. APMIS 2011;119:330-5. [PubMed]
- Fisher DE, Barnhill R, Hodi FS, et al. Melanoma from bench to bedside: meeting report from the 6th international melanoma congress. Pigment Cell Melanoma Res 2010;23:14-26.
- Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med 2010;363:2191-9. [PubMed]
- Cheli Y, Ohanna M, Ballotti R, et al. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 2010;23:27-40. [PubMed]
- Hou L, Pavan WJ. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: do all roads lead to Mitf? Cell Res 2008;18:1163-76. [PubMed]
- Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005;436:117-22. [PubMed]
- Wellbrock C, Rana S, Paterson H, et al. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PLoS One 2008;3:e2734. [PubMed]
- Wu M, Hemesath TJ, Takemoto CM, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev 2000;14:301-12. [PubMed]


