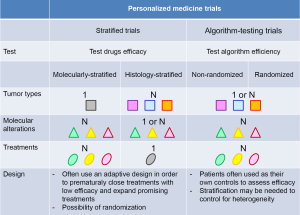
The spectrum of clinical trials aiming at personalizing medicine
Introduction
Personalized medicine, also called precision medicine, is defined by the National Cancer Institute as “A form of medicine that uses information about a person’s genes, proteins, and environment to prevent, diagnose, and treat disease”. This term emerged with molecularly targeted agents 15 years ago. While cytotoxic agents destroy rapidly dividing cells by triggering DNA and cell division machinery, molecularly targeted agents block a peculiar molecular alteration involved in cell proliferation, angiogenesis, metastasis, invasion, etc. Medical indication of around half of the drugs approved for clinical use integrates the presence of a molecular alteration (1). The development of some of these agents in molecularly-defined subgroups of patients has yielded unprecedented efficacy in some tumor types (2-7). The remaining drugs lack a validated predictive biomarker of efficacy, and include for example anti-angiogenic agents or mammalian target of rapamycin (mTOR) inhibitors.
Molecularly targeted agents have consistently followed the same clinical development as cytotoxic agents based on tumor location and histology, although some molecular alterations have been reported across different tumor types (8). The emergence of molecularly targeted agents has not immediately led to a paradigm shift in drug development for the following reasons: (I) molecular alterations were initially thought to be specific of certain tumor types, such as the BCR/ABL fusion gene in chronic myeloid leukemia; (II) the functional significance of some molecular alterations varies across tumor types, as illustrated by the limited efficacy of BRAF inhibitors in BRAF V600E-mutated colorectal cancer (9) and the substantial efficacy of these drugs in BRAF V600E-mutated melanoma (3); (III) histology-independent drug development would be challenged by the lack of valid benchmarks represented by data on drug efficacy in patients with any type of cancer harboring a common molecular alteration.
Since recently, advances in high-throughput technologies have allowed depicting most druggable molecular alterations for an affordable cost in a timeframe compatible with clinical practice. Despite the caveats associated with histology-independent drug development mentioned above, the question whether personalized medicine based on the molecular profiling of the tumor of cancer patients would still improve their outcome has arisen and led to set up studies addressing this question. We identified three distinct types of studies aiming at personalizing medicine, including molecular screening programs using molecular profiling of the tumor, as well as two distinct types of personalized medicine trials: (I) stratified clinical trials that can be stratified according to either molecular alterations or tumor types; and (II) algorithm-testing trials that evaluate a treatment algorithm instead of drugs’ efficacy. These studies are associated with numerous challenges that are then discussed.
Molecular screening programs
Several molecular screening programs have been set up around the world (Table 1). These screening programs seek genomic molecular alterations including DNA mutations and/or gene copy number alterations. The primary objective behind these screening programs is to guide patients to clinical trials evaluating drugs matching identified molecular alterations. Most of these programs have therefore been offered to patients with recurrent and/or metastatic cancer with the aim of better selecting therapy in the absence of standard of care. However, some of the programs are proposed to patients at any stage of their disease. These latter programs might be very costly and restrictions might be needed by targeting specific patient populations. Some of the programs focus on one specific tumor types such as non-small cell lung cancer (NSCLC) and breast cancer, while the remaining majority are opened to patients with any kind of cancer.
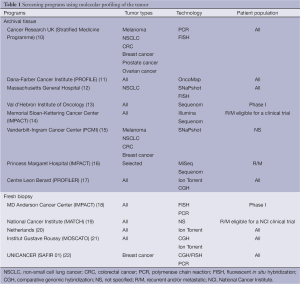
Full table
Various technologies are used in these screening programs, most of them relying on high throughput technologies, while a few programs only look at a very limited number of molecular alterations using polymerase chain reaction (PCR) and fluorescent in situ hybridization (FISH). The use of high-throughput technologies often addressed additional challenges based on the complexity and the size of the data. In practice, the management and the analysis of such data require adequate bioinformatics environment and resources.
In the majority of the screening programs, molecular analyses are proposed on archival tissue from primary tumor with the underlying assumption that molecular alterations identified on the primary tumor would still be relevant at later stages of the disease. Numerous studies have evaluated the concordance rate between primary and metastatic sites in terms of actionable molecular alterations (23). Results of these studies are inconsistent, with discrepancies rates reported being low in colorectal (24) and lung cancer (25) for instance and much higher ones in breast cancer (26,27). Discrepancies may account for several factors, including tumor type, type of molecular alteration and most importantly selection pressure due to molecularly targeted therapy, but also the percentage of tumor cells, tumor heterogeneity and the emergence of sub-clones potentially related to the targeted therapy. As an example, epidermal growth factor receptor (EGFR)-mutated lung cancer patients might ultimately become resistant to EGFR-targeting therapy through the emergence of the T790M EGFR mutation (28). Screening programs based on the identification of molecular alterations on archival tissue are easier to implement than those mandating a tumor sample from a metastatic site. The question remains whether the molecular data collected from the primary tumor is adequate or rather expose patients to be guided to inadequate therapy.
These screening programs provide useful information on the prevalence of specific molecular alterations in various tumor types. They might also identify predictive biomarkers as well as new molecular alterations although this should become infrequent because of the data accumulated by the genome sequencing of hundreds of tumors that are now available. Besides the cognitive aspect, these screening programs might have a direct implication for the patients by allowing them to potentially use the information from their tumor for further therapy. Most of the programs include a retrospective assessment of the efficacy in their objectives. Retrospective analysis of the outcome of cancer patients included in a phase I trial at the M.D. Anderson Cancer Center based on a molecular alteration identified on metastatic tumor samples showed that it was better than the outcome of patients who entered a phase I trial without matching in terms of overall response rate, failure-free and overall survival (18). The ratio of progression-free survival (PFS) on phase I therapy to PFS on last therapy was also substantially longer in the former group of patients. Results in terms of PFS ratio were reproduced in the similar MOSCATO study (21). In contrast, disappointing results were reported in a subgroup of patients with metastatic colorectal cancer referred to phase I trials at the Val d’Hebron Hospital (29).
Overall, these retrospective analyses of the utility of screening programs suggest that guiding patients to molecularly targeted therapy would improve their outcome. This has constituted the rationale for designing prospective personalized medicine clinical trials, including stratified and algorithm-testing personalized medicine trials (Figure 1). The former ones evaluate the efficacy of drugs while the latter ones evaluate the relevance of a treatment algorithm.
Stratified personalized medicine trials
Stratified personalized medicine trials include two distinct types of trials: molecularly- and histology-stratified trials (Table 2). Molecularly-stratified trials usually evaluate several drugs but focus on one specific tumor type, while histology-stratified trials, also called basket trials, focus on one specific drug across diverse tumor types.
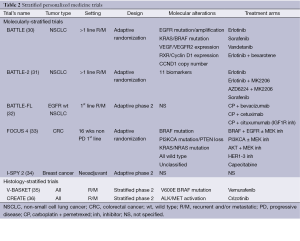
Full table
Molecularly-stratified trials
Molecularly-stratified personalized medicine trials allocate drugs or drug combinations to patients based on the presence or the absence of specific molecular alterations. These trials sometimes use adaptive designs so that arms with little efficacy can be closed early, whereas arms that show hints of efficacy can be expanded.
The BATTLE trial is the first molecularly-stratified adaptive randomized clinical trial (30). The objective of this trial was to test several biomarkers together with several drugs and to find out what were the most successful associations in patients with advanced lung adenocarcinoma. Patients were randomized between four treatments arms: erlotinib, vandetanib, erlotinib plus bexarotene, and sorafenib. All patients included in the trial were tested for the following molecular alterations on a tumor sample from a mandatory biopsy: EGFR mutation/amplification, KRAS/BRAF mutation, VEGF/VGFR2 expression, RXR/Cyclin D1 expression and CCND1 copy number. Real-time analysis of efficacy using a Bayesian model led to further evaluation of the efficacy of sorafenib in patients whose tumor harbored a KRAS mutation. The BATTLE-2 trial (NCT01248247) allocates advanced lung cancer patients who progress on first line chemotherapy to one of the four following drugs or drug combinations based on the analysis of 11 biomarkers: erlotinib, erlotinib in combination with MK2206 (AKT inhibitor), MK2206 in combination with AZD6224 (MEK inhibitor), and sorafenib (31). The trial plans to accrue 450 patients. BATTLE-FL (front-line) (NCT01263782) allocates treatment-naïve metastatic and/or recurrent EGFR wild-type lung cancer patients to one of the three following drugs in combination with the doublet chemotherapy carboplatin and pemetrexed based on the molecular profile established on a mandatory biopsy of a metastatic site: bevacizumab, cetuximab and cituxumumab (anti-IGF1R inhibitor) (32). The BATTLE-2 and BATTLE-FL trials are ongoing.
The FOCUS 4 is a phase II/III trial that involves metastatic colorectal patients without progression after first-line therapy at 16 weeks (33). Following molecular alterations identified on a tumor sample from a metastatic site, patients are allocated to one of the five maintenance treatment arms depending on the molecular alterations identified: (I) a combination of a BRAF inhibitor, an EGFR inhibitor with or without a MEK inhibitor in case of KRAS, BRAF, or NRAS mutation; (II) a PI3KCA inhibitor with or without a MEK inhibitor in case of PI3KCA mutation; (III) a combination of an AKT inhibitor and a MEK inhibitor in case of PTEN loss; (IV) a pan-HER inhibitor if wild type for all previous molecular alterations; and (V) capecitabine if unclassified. Twenty four hundred patients will be included with the aim of randomizing 1,536 patients. In each treatment arm, patients will be randomized against placebo with a 2:1 ratio. An adaptive design is used so that promising treatment arms can switch from a phase II to a phase III. A substantial advantage of this trial is its ability to include any colorectal cancer patient as molecular stratification covers all molecular subgroups.
The I-SPY 1 study had identified clinical, imaging and genomic predictive markers of pathological complete response based on the analysis of 221 early breast cancer patients treated with neoadjuvant chemotherapy (37). Based on the hypotheses generated by this study, the I-SPY 2 trial (NCT01042379) has been then set up in the same patient population (34). Patients with stage 3 breast cancer are randomized between standard neoadjuvant chemotherapy and the same treatment combined with a molecularly targeted agent based on molecular alterations identified on tumor biopsy. The primary endpoint is the pathological complete response rate. The protocol allows opening new treatment arms during the trial, to early close presumably non-efficient treatment arms and to expand promising arms. A Bayesian framework is used meaning that the error rate is not controlled according to usual standards. Randomization allows comparing the efficacy of the different experimental arms to standard of care.
All these molecularly-stratified trials evaluate different treatment strategies in different tumor types and settings. However, they do not provide the same level of evidence as 2-arms randomized trials. To date, the operating characteristics of most of these designs have been evaluated in a limited number of settings. As opposed to molecularly-stratified trials, histology-stratified or basket trials evaluate the efficacy of one drug in multiple tumor types based on the presence or the absence of specific molecular alterations, usually matching the targets of the drug under evaluation.
Histology-stratified trials
The V-BASKET trial (NCT01524978) seeks for signals of efficacy in recurrent and/or metastatic cancer patients whose tumors harbor a BRAF mutation, except in patients with V600E BRAF-mutated melanoma (35). The CREATE trial (NCT01524926) is a similar trial with crizotinib intended for the same patient population whose tumors harbor a ALK or MET molecular alteration except for ALK-translocation in lung adenocarcinoma (36). Both trials allow the molecular analyses to be performed on archival tissue, therefore offering the possibility to perform the molecular analyses while the patient is on another treatment. The caveat of this strategy is prescribing a drug in the recurrent and/or metastatic setting based on a molecular alteration detected on the primary tumor.
The main drawbacks of these histology-stratified trials are that (I) they may require to screen many patients who will not be treated with the matching targeted therapy if the incidence of the molecular alterations is low, except if they are concomitantly included in a screening program; and (II) they are not randomized that means, the activity is compared to some theoretical value that is very difficult to interpret due to the variety of tumor types that are included. In all these trials, sample size is calculated so that the efficacy of a drug or drug combination can be adequately assessed with a pre-specified power in any type of cancer.
Algorithm-testing personalized medicine trials
Clinical trials that include patients with multiple tumor types and in which multiple molecular alterations are tested for further treatment allocation can formally evaluate the efficacy of a specific drug (or drug combination) in a specific molecularly-characterized subgroup of patients with a same tumor type only if (I) the trial is stratified on all subgroups of patients; and (II) a formal sample size has been calculated in each subgroup for each drug (or drug combination) to get enough power to conclude based on pre-specified types I and II errors. In other words, except if the sample size is huge, results from such trials will not allow drawing any robust conclusion regarding the potential efficacy of a specific drug or drug combination in a given molecularly-defined subgroup of patients. These trials in fact can only evaluate the treatment algorithm that has been set up to allocate treatments to patients, regardless of the treatment administered. One fundamental requirement in these trials is that the treatment algorithm is not modified during the study. Algorithm-testing trials include non-randomized trials that usually use patients as their own control to assess efficacy, and randomized trials that address various questions (Table 3).
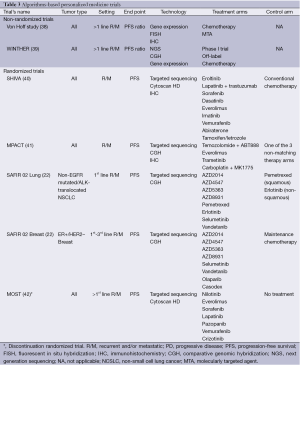
Full table
Non-randomized clinical trials
Von Hoff’s study is the first published histology-independent clinical trial using tumor molecular alterations to select treatment (38). Patients with any type of recurrent and/or metastatic cancer that was refractory to standard of care had selected molecular alterations analyzed using immunohistochemistry (IHC), FISH and oligonucleotide microarray gene expression assays. Based on the detected molecular alterations, a drug or drug combination was prescribed. Eighteen of the 66 treated patients (27%) had a ratio of the time to progression (TTP) on matching targeted treatment to the TTP under the last previous treatment >1.3, which was statistically different from the hypothesis that this ratio is one in the absence of treatment effect.
More recently, the WIN consortium launched the WINTHER trial (NCT01856296) that is currently open in several countries (39). This trial is open to patients with any kind of refractory advanced cancer. Two samples are taken from the patient, one from a metastatic site and the second from adjacent normal tissue. Druggable molecular alterations are first investigated from the tumor sample using next generation sequencing for mutations screening and CGH array for gene copy number alterations profiling. If a druggable molecular alteration is identified, patients are either guided to a phase I clinical trial with an agent presumably matching the molecular alteration or are being prescribed an already approved molecularly targeted agent off-label. If no druggable molecular alteration is detected, data from tumoral RNA and RNA from normal adjacent tissue are analyzed in order to identify gene expression profiles that can orient the patient to the best therapy. Both treatment arms will be analyzed separately using PFS ratio as a primary endpoint. It is planned to treat 200 patients, 80 in the former arm and 120 the latter one, based on the assumption that a druggable molecular alteration will be present in 40% of patients. The main advantage of this trial is that all included patients will be treated.
Given the multiple tumor types included in these non-randomized trials, one way to evaluate treatment efficacy has been to use patients as their own controls assessing the PFS ratio. The main criticism to the use of the PFS/TTP ratio as a primary endpoint in these studies is the assessment of PFS/TTP on the last therapy outside of the clinical trial. In addition, the underlying assumption behind this endpoint is that the natural history of disease is linear over time, in other words that the two PFS/TTP are correlated, which might not be true. For these reasons, the use of randomization has been suggested (43).
Randomized clinical trials
Algorithm-testing randomized clinical trials have been set up either for all types of cancers or in specific tumor types.
The SHIVA trial (NCT01771458) is a proof-of-concept randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional chemotherapy in patients with any type of cancer that is refractory to standard of care (40). The primary endpoint is PFS. The trial is stratified on (I) the patient’s prognosis using the Royal Marsden Hospital prognostic score for phase I cancer patients (44); and (II) the signaling pathway the selected molecular alteration belongs to. Molecular alterations are evaluated on a tumor sample from a metastatic site using the AmpliSeq Cancer panel on Ion Torrent sequencing (Life Technologies) for mutations screening, the Cytoscan HD technology (Affymetrix®) for gene copy number alterations profiling, and IHC for estrogen, progesterone and androgen receptors expression analyses. Only marketed molecularly targeted agents are used in this trial according to a pre-specified treatment algorithm. Eleven molecularly targeted agents are available within the clinical trial, whereas conventional chemotherapy is prescribed at the physician’s discretion in the control arm. Cross-over is proposed in both arms at disease progression, allowing the evaluation of tumor growth kinetics on both treatments for each patient (45). Physicians are being told the molecular alteration of interest for their patient only at the time they are about to be treated in the experimental arm. The randomization of 200 patients is planned to detect a significant difference in 6-month PFS with a bilateral type I error of 5% and an 80% power. Feasibility results on the first 100 included patients have shown that biopsies are safe and that 40% of patients were detected a molecular alteration that allowed them to be randomized (46). Ancillary studies include the evaluation of the ability of circulating DNA to predict treatment efficacy or resistance, as well as a medico-economic evaluation of the experimental strategy. Efficacy results should be available in 2016.
The MPACT trial is a randomized phase II trial that will include the same patient population asin the SHIVA trial (41). A tumor sample of a metastatic site will also be mandatory. Molecular alterations will be detected using the Ion Torrent sequencing (Life Technologies) for mutations screening, CGH array (Agilent) for gene copy number alterations profiling, and IHC for protein expression evaluation. Patients will be randomized between therapy matching the detected molecular alteration and therapy not matching the detected molecular alteration. Cross-over will be proposed at disease progression for patients randomized in the non-matching treatment arm. Sample size calculation based on the results of Tsimberidou et al., with an expected overall response rate of 25% in the matching treatment arm versus 5% in the non-matching treatment arm (18). The randomization of 200 patients is planned. Although it might be difficult for patients to accept the randomization in the non-matching treatment arm, this design is the only one that evaluates solely the treatment algorithm. Accrual should start in 2014.
The MOST trial (EudraCT: 2012-004510-34) is a randomized discontinuation trial for patients who have progressed on first-line treatment for a recurrent and/or metastatic cancer (42). Molecular alterations will be identified on a sample from either a metastatic site or the primary using the Ion Torrent technology (Life Technologies) for mutations screening and CGH array (Agilent) for gene copy number alterations profiling. Patients will be treated during three months according to a pre-specified algorithm with one of the seven available already marketed molecularly targeted agents. Responding patients will continue on therapy, while progressive patients will be taken off study. Patients with stable disease will be randomized between treatment continuation and discontinuation for three months. The MOST trial will provide a more accurate evaluation of efficacy than a single-arm study by deciphering between disease stabilization related to the natural history of the disease and disease stabilization related to a cytostatic effect of molecularly targeted therapy. The trial has recently opened.
The SAFIR 02 trials are tumor-specific randomized trials evaluating maintenance therapy (22). SAFIR 02 Breast will include patients with HER-2 negative and estrogen receptor positive recurrent and/or metastatic breast cancer who have not progressed after four to eight cycles of first- to third-line chemotherapy, while SAFIR 02 Lung will include patients with EGFR and ALK wild types recurrent and/or metastatic lung cancer who have not progressed after four cycles of first-line platinum-based chemotherapy. All patients will have a tumor sample taken from a metastatic site in order to seek molecular alterations using targeted sequencing for mutations analysis and CGH array for gene copy number alterations analysis. Patients will be randomized between a molecularly targeted agent from AstraZeneca matching the detected molecular alteration and maintenance chemotherapy (pemetrexed for squamous lung cancer and erlotinib for non-squamous lung cancer). The primary endpoint will be PFS. These trials have opened in 2014 as well. These trials evaluate the utility of a treatment algorithm for selecting maintenance therapy following chemotherapy in recurrent and/or metastatic luminal breast cancer and lung cancer not eligible for molecularly targeted therapy.
All these algorithm-testing clinical trials base their algorithm on DNA analysis, except the WINTHER trial that analyses also gene expression in case no molecular alteration has been detected on DNA. These trials include only so far patients with recurrent and/or metastatic cancer. All these trials ultimately address with different angles the question of whether the use of tumor molecular profiling would improve the outcome of these patients. Results of these trials are highly expected as they will provide meaningful information as of high-throughput technologies should or should not be used in routine in the future. None of these trials is powered to adequately assess the efficacy of any treatment in any molecularly-defined subgroup of patients with a same tumor type and histology. Except for the MPACT trial (41), the treatment effect is confounded with the treatment algorithm for treatment allocation. If a targeted agent had a tremendous effect regardless of the molecular alteration, the whole arm (or period in trials using the PFS/TTP ratio) would appear to be superior and one might erroneously conclude that selecting the treatment based on the molecular profile is superior to not using the treatment algorithm.
Challenges
Given their complexity, personalized medicine trials are associated with numerous challenges. These trials indeed involve several different crucial stakeholders, including physicians, radiologists, pathologists, biostatisticians, sequencing platforms managers, bioinformaticians and biologists. While the four former ones had been used to work together in a clinical setting, the latter ones usually work with researchers and are not used time constraints related to patients care. The novelty with personalized medicine trials is that all these people have to coordinate their actions so that treatment decisions for cancer patients are timely taken.
Tumor tissue
Many clinical trials require having a tumor sample taken from a metastatic site. While biopsies of metastatic sites have been long shown to be feasible without excessive complications (46), a metastatic site might not always be easily accessible for sampling. Moreover, some metastatic sites might still be accessible but not appropriate for high- throughput technologies such as bone where the sequencing failure rate is high (22). Developments of less invasive and more convenient procedures are highly expected. In addition, these samples cannot appreciate tumor heterogeneity.
Most of developments have focused so far on liquid biopsies, including circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA). The prerequisite to use this material is obviously to be able to detect it in patients’ blood, which might not always be the case. While it has been reported that specific molecular alterations can be detected in CTCs or ctDNA, it remains to be demonstrated whether high-throughput technologies can be successfully applied on such material. These liquid biopsies also present a priceless advantage of allowing sequential sampling with the premise of identifying pharmacodynamic biomarkers of efficacy as well as resistance biomarkers. In addition, they might theoretically be able to appreciate tumor heterogeneity if circulating tumor material reflects the tumor burden.
Technology
The use of high-throughput technologies is a multistep process necessitating in the clinics the interaction between multiple actors in order to get real time results for treatment decision making. These steps include tumor sampling by surgeons or interventional radiologists, histological diagnostic confirmation by pathologists, DNA extraction, sequencing, bioinformatics analyses and biological validation by biologists. All these steps are associated with error margins (47), and the whole multistep process might as a consequence be associated with an even increased error margin. Quality controls must therefore be performed at each step of the process and traceability is required.
While the implementation of high-throughput technologies in clinical trials is challenging, the techniques per se represent another challenge. First, the choice of the techniques, including DNA extraction kits, sequencers, pipelines, bioinformatics pipelines must be carefully decided, so that the rates of false negatives and positives reach a reasonable threshold in regards to the project. Different sequencers might also produce different results, although discrepancies are infrequent (48). Second, the techniques have to be validated before being implemented in any research program. No recommendations exist to date regarding these validations. Do we need to validate the technology and some case control somatic mutations by Sanger sequencing in a pilot study before starting any program or should we validate any detected mutation during the program? Finally, techniques evolve so quickly that one might not have any other choice than to implement changes during a trial, simply because the technique used initially is not available any more. In any case, techniques changes must be precisely described when results of clinical trials are reported. The National Cancer Institute, in collaboration with scientists representing multiple areas of expertise relevant to ‘omics’-based test development, has developed a checklist of criteria that can be used to determine the readiness of omics-based tests for guiding patient care in clinical trials (49).
Biology
Behind all these new technologies that are implemented in clinical trials, the real challenge relies on the biological assumptions made in the treatment algorithms used. The elaboration of treatment algorithms have to be supported by strong biological hypotheses. The biological assumptions can either be based on clinical data or on preclinical data only. The key question is the choice of the level of evidence required to set up a treatment algorithm. Is clinical efficacy demonstrated with trastuzumab in HER-2-amplified breast and gastric cancer patients enough to justify the use of this drug in any HER-2-amplified cancer patients? Is the preclinical description of an activating molecular alteration in one single paper enough to incorporate it in the algorithm?
It is now clear that the biological significance of some molecular alterations varies depending on the tumor types, such as the BRAF V600E mutation. Most of melanoma patients respond to BRAF inhibition whereas only a low proportion of colorectal cancer patients do. Preclinical data have shown that colorectal cancer cells may escape via a feedback loop involving EGFR, which is often expressed in colorectal cancer as opposed to melanoma (9). The differential biological significance observed for a specific alteration might indeed be explained by other molecular alterations. Taking into account multiple molecular alterations (or modulations) to predict treatment efficacy opens the field of “systems biology”. The “systems biology” approach will need considerable bioinformatics research to produce valid tools to be used in the clinic. It remains to be demonstrated that a systems biology approach can improve the patients’ outcome. It will also surely imply that several pathways are implicated which raises the critical question of drug combinations that are not easy to manipulate. To date, only single molecular alterations are used to drive treatment selection. No multidimensional algorithm has been proved to be superior.
Another issue pertains when several druggable molecular alterations are detected. The ideal solution would be to target all of them with matching drugs. However, drugs are not easy to combine because of their often overlapping toxicities (50). Treatment priorities have to be established based on strong preclinical data whenever possible. Sequential use of drugs would be worth evaluating especially if pharmacodynamic markers are easily assessable.
New preclinical data and clinical case reports are published every week and improve our knowledge of cancer biology. However, from a clinical research perspective, it is essential that the treatment algorithm does not change all along the trial for obvious reproducibility purposes. It is reasonable however considering an enrichment of the algorithm if it does not question previous treatment decisions.
Statistics
Personalized medicine trials that might involve several tumor types, several molecular alterations and matching treatments represent a new challenge for biostatisticians. The ideal clinical trial design would determine a sample size so that a treatment effect can be evaluated with enough statistical power in any subgroup of patients with a specific tumor type harboring a specific molecular alteration and treated with a specific treatment. This type of clinical trial would obviously require thousands of patients and would not be feasible in practice. Compromises have therefore to be made, including stratification in randomized trials that allows controlling for heterogeneity or taking patients as their own controls in non-randomized trials.
Costs
Although the costs of high-throughput technologies are decreasing exponentially, the overall cost of personalized medicine trials remains high. These costs include the sequencing per se, but also the associated bioinformatics analyses and data storage. Trials using drugs that are already on the market should preferentially have the drugs funded by the companies. This might be tricky to get agreements with pharmaceutical companies if several drugs are used, especially if the trial is not powered to evaluate the efficacy of the drug which is less appealing from a pharmaceutical company’s point of view. In the case companies refuse to provide their drugs, drugs have then to be funded by the sponsor, which might substantially increase the budget of the trial, especially when patients fortunately display prolonged tumor responses. If studies eventually demonstrate that the use of high-throughput technologies improves patients’ outcome, the cost of the implementation of these technologies in routine will have to be precisely determined. The implementation of high-throughput technologies may need a complete restructuration of hospitals unless these analyses are outsourced. In any case, discussions with health authorities but also with pharmaceutical companies will have to be engaged to discuss cost-sharing. Pharmaceutical companies may indeed derive benefits from such implementations that allow patients to be guided in specific molecular-based clinical trials. Medico-economic analyses associated to personalized clinical trials are in that sense crucial.
Ethics
Personalized medicine trials rise ethical considerations if constitutional DNA is needed for genomic analyses (e.g., for exome sequencing). Once constitutional DNA of a given patient is available, following questions have to be answered: Are there genomic data that have to be investigated? What information should be brought back to the patients? What if specific information that concerns the descendants becomes available after the patient’s death? It is therefore essential that consent forms used in personalized medicine trials precisely anticipate these questions. They should preferably be discussed with patients’ advocates.
Drugs
Access to drugs represents an important issue in personalized medicine trials, especially when several drugs are used. As mentioned earlier, pharmaceutical companies may not be very keen to provide their drugs in multi-drugs trials, all the more if the drugs are in clinical development. Drug combinations with drugs from different companies are almost impossible to obtain, while obtaining drugs from a same company for a drug combination still remains challenging. In addition, algorithm-testing personalized medicine clinical trials do not directly benefit pharmaceutical companies, since these trials evaluate treatment algorithms and not drugs’ efficacy. However, they might still derive indirect benefits by the use of systematic molecular profiling of patients that might improve the inclusion rate in clinical trials based on specific molecular alterations. It is therefore urgent that pharmaceutical companies adhere to such trials, as AstraZeneca in the SAFIR 02 trial that provided part of its pipeline.
Discussion
The emergence of cytotoxic chemotherapy after the Second World War has led to a significant improvement in cancer cure. While molecularly targeted therapy has clearly modified the prognosis of some cancers such as chronic myeloid leukemia or subgroups of cancers such as HER-2-overexpressing breast cancer, the cure rates of cancer patients has not increased substantially. Two reasons may explain this. First, a minority of cancer patients is today eligible for molecularly targeted therapy. Second, molecularly targeted therapy is mostly approved in the recurrent and/or metastatic setting where they prolong survival but do not cure. Only trastuzumab in HER-2-overexpressing breast cancer and imatinib in c-KIT-overexpressing gastrointestinal stromal tumors are approved in the adjuvant setting (51,52). The substantial decrease of recurrences in these two settings likely provides an indirect demonstration that these two agents are able to cure cancer. The fundamental question we have today is whether the use of high-throughput technologies will increase the rate of cancer cure. The personalized medicine trials described above are almost all performed in patients with recurrent and/or metastatic cancer and will certainly not lead to an increased cure rate of cancer even if they are positive. Only the evaluation of such strategies at earlier stages of the disease could potentially lead to substantially improve the rate of cancer cure. While positive results of these trials would undoubtedly accelerate the implementation of high-throughput technologies in routine, negative results should not be interpreted as a failure of the overall strategy. Subgroup analyses might also pinpoint potential biomarkers that might after clinical validation improve treatment efficacy when taken into account. Patients are indeed usually heavily pretreated in these trials. In addition, patients are usually proposed single agent molecularly targeted therapy, which we know is often insufficient to achieve prolonged efficacy. Last, the treatment algorithms used in these trials have not been validated. Bioinformatics and research in biology will be critical to improve them, using systems biology approaches, along with functional validation in preclinical studies.
The personalized medicine trials described above focus on the use of genomic alterations to decide molecularly targeted therapy. Other approaches to treat cancer have recently emerged and appear to be very promising, including immunotherapy and therapies targeting the microenvironment. Restoring an efficient immune response by targeting CTLA4 in melanoma patients has been demonstrated to improve their outcome in the recurrent and/or metastatic setting (53). Ten to fifteen percent of patients are long responders to this treatment, which is unprecedented. The future will tell whether they are cured, which might be plausible given the mechanism of action of these drugs. Outstanding results have also been reported in several tumor types with drugs targeting the PD-L1/PD1 axis (54-56). Other drugs that target the microenvironment such anti-CSF-1R antibodies that target activated macrophages on the surface of which CSF-1R is present are in clinical development.
Ultimately, it is very likely that cure of cancer will substantially increase thanks to a combination of molecularly targeted therapy that will be more adequately implemented using high-throughput technologies and novel therapies such as immunotherapy and therapies targeting the microenvironment. The integration of these latter therapies to molecularly targeted therapy opens an important field in cancer research. The development of ctDNA will hopefully also help circumvent the issue of intra-tumor heterogeneity (57).
Acknowledgements
This work was supported by a grant from the Institut National du Cancer (#2010-1-PL SHS-06-IC-1).
Disclosure: The authors declare no conflict of interest.
References
- Sleijfer S, Ballman K, Verweij J. The future of drug development? Seeking evidence of activity of novel drugs in small groups of patients. J Clin Oncol 2013;31:2246-8. [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [PubMed]
- Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012;366:707-14. [PubMed]
- Druker BJ, Talpaz M, Resta DJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med 2001;344:1031-7. [PubMed]
- Sekulic A, Migden MR, Oro AE, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 2012;366:2171-9. [PubMed]
- Slamon D, Eiermann W, Robert N, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med 2011;365:1273-83. [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [PubMed]
- Ciriello G, Miller ML, Aksoy BA, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet 2013;45:1127-33. [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [PubMed]
- Walker I, MacDonald F, Stuart H, et al. Utilizing a collaborative working model to optimize molecular analysis of solid tumors in the Cancer Research UK’s Stratified Medicine Programme. J Clin Oncol 2013;31 (suppl; abstr 11094).
- Rollins BJ, MacConaill LE, Wagle N, et al. PROFILE: Broadly based genomic testing for all patients at a major cancer center. J Clin Oncol 2013;31 (suppl; abstr 1531).
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [PubMed]
- Rodón J, Saura C, Dienstmann R, et al. Molecular prescreening to select patient population in early clinical trials. Nat Rev Clin Oncol 2012;9:359-66. [PubMed]
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=745680&version=HealthProfessional&protocolsearchid=12174081
- Available online: www.vicc.org
- Bedard PL, Oza AM, Tsao MS, et al. Princess Margaret Cancer Centre (PMCC) Integrated Molecular Profiling in Advanced Cancers Trial (IMPACT) using genotyping and targeted next-generation sequencing (NGS). J Clin Oncol 2013;31 (suppl; abstr 11002).
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=745671&version=HealthProfessional&protocolsearchid=12176142
- Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The M. D. Anderson Cancer Center Initiative. Clin Cancer Res 2012;18:6373-83. [PubMed]
- Kaiser J. Rare cancer successes spawn ‘exceptional’ research efforts. Science 2013;340:263. [PubMed]
- Available online: http://www.eacr.org/mpathcourse2012/presentations/E%20Voest_presentation.pdf
- Hollebecque A, Massard C, De Baere T, et al. Molecular screening for cancer treatment optimization (MOSCATO 01): A prospective molecular triage trial—Interim results. J Clin Oncol 2013;31 (suppl; abstr 2512).
- André F, Bachelot T, Commo F, et al. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER). Lancet Oncol 2014;15:267-74. [PubMed]
- Vignot S, Besse B, André F, et al. Discrepancies between primary tumor and metastasis: a literature review on clinically established biomarkers. Crit Rev Oncol Hematol 2012;84:301-13. [PubMed]
- Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012;30:2956-2962. [PubMed]
- Vignot S, Frampton GM, Soria JC, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol 2013;31:2167-72. [PubMed]
- Amir E, Miller N, Geddie W, et al. Prospective study evaluating the impact of tissue confirmation of metastatic disease in patients with breast cancer. J Clin Oncol 2012;30:587-92. [PubMed]
- Dupont Jensen J, Laenkholm AV, Knoop A, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res 2011;17:667-77. [PubMed]
- Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet 2005;37:1315-6. [PubMed]
- Dienstmann R, Serpico D, Rodon J, et al. Molecular profiling of patients with colorectal cancer and matched targeted therapy in phase I clinical trials. Mol Cancer Ther 2012;11:2062-71. [PubMed]
- Kim ES, Herbst RS, Wistuba II, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov 2011;1:44-53. [PubMed]
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=690229&version=HealthProfessional&protocolsearchid=12174127
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=691870&version=HealthProfessional&protocolsearchid=12174129
- Kaplan R, Maughan T, Crook A, et al. Evaluating many treatments and biomarkers in oncology: a new design. J Clin Oncol 2013;31:4562-8. [PubMed]
- Barker AD, Sigman CC, Kelloff GJ, et al. -SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther 2009;86:97-100. [PubMed]
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=724784&version=HealthProfessional&protocolsearchid=12174135
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=724783&version=HealthProfessional&protocolsearchid=12174139
- Esserman LJ, Berry DA, DeMichele A, et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL--CALGB 150007/150012, ACRIN 6657. J Clin Oncol 2012;30:3242-9. [PubMed]
- Von Hoff DD, Stephenson JJ Jr, Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28:4877-83. [PubMed]
- Available online: http://www.cancer.gov/clinicaltrials/search/view?cdrid=749710&version=HealthProfessional&protocolsearchid=12176033
- Le Tourneau C, Kamal M, Trédan O, et al. Designs and challenges for personalized medicine studies in oncology: focus on the SHIVA trial. Target Oncol 2012;7:253-65. [PubMed]
- Kummar S. Molecular Profiling based Assignment of Cancer Therapy (MPACT). Proc NCI-EORT-ASCO meeting, 2013.
- Available online: http://www.cancer-lyric.com/programme/programme-1/
- Doroshow JH. Selecting systemic cancer therapy one patient at a time: is there a role for molecular profiling of individual patients with advanced solid tumors? J Clin Oncol 2010;28:4869-71. [PubMed]
- Arkenau HT, Barriuso J, Olmos D, et al. Prospective validation of a prognostic score to improve patient selection for oncology phase I trials. J Clin Oncol 2009;27:2692-6. [PubMed]
- Le Tourneau C, Servois V, Diéras V, et al. Tumour growth kinetics assessment: added value to RECIST in cancer patients treated with molecularly targeted agents. Br J Cancer 2012;106:854-7. [PubMed]
- Le Tourneau C, Paoletti X, Servant N, et al. Randomised proof-of-concept phase II trial comparing targeted therapy based on tumour molecular profiling vs conventional therapy in patients with refractory cancer: results of the feasibility part of the SHIVA trial. Br J Cancer 2014. [Epub ahead of print]. [PubMed]
- O’Rawe J, Jiang T, Sun G, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med 2013;5:28. [PubMed]
- Ratan A, Miller W, Guillory J, et al. Comparison of sequencing platforms for single nucleotide variant calls in a human sample. PLoS One 2013;8:e55089. [PubMed]
- McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials. Nature 2013;502:317-20. [PubMed]
- Soria JC, Massard C, Izzedine H. From theoretical synergy to clinical supra-additive toxicity. J Clin Oncol 2009;27:1359-61. [PubMed]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 2005;353:1659-72. [PubMed]
- Dematteo RP, Ballman KV, Antonescu CR, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 2009;373:1097-104. [PubMed]
- Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711-23. [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [PubMed]
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [PubMed]
- Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013;369:134-44. [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [PubMed]