
Moving toward multi-dimensional biomarkers in cancer immunotherapy
The development of anti-programmed death 1 (PD1) antibodies has rapidly changed standard oncology clinical practice however only a fraction of patients benefit from these treatments and some tumor types remain unresponsive. Importantly then, biomarker development for the appropriate selection of patients and to inform novel therapeutic development is essential in the field of cancer immunotherapy. Ott et al. recently reported on the analysis of KEYNOTE-028, which was a non-randomized, phase 1b basket trial that enrolled 475 patients with advanced programmed death ligand 1 (PD-L1) positive solid tumors of multiple histologies, and treated with pembrolizumab 10 mg/kg every 2 weeks for up to 2 years (1). The primary end point was objective response rate with secondary endpoints surrounding safety and other survival analyses. The study reported a range of response rates across tumor types from 0% in pancreatic cancer up to 33% in small-cell lung cancer, investigating over 20 cancer types in total. Beyond clinical outcomes, Ott et al. examined relationships between T cell-inflamed gene expression profile (GEP), PD-L1 expression, and tumor mutational burden (TMB) as well as antitumor activity in relationship to outcomes and between biomarkers. Correlations were observed between markers of T cell-inflamed tumors, both GEP or PD-L1 immunohistochemistry, and response to pembrolizumab as well as an independent correlation between TMB and response, suggesting that multiple mechanisms may facilitate response to checkpoint immunotherapy.
These biomarker observations made in KEYNOTE-028 build on years of basic and translational immunology research which are of urgent relevance again as the field considers optimal patient selection and perhaps next generation approaches for cancer immunotherapy. Seminal associations of overall survival and CD8 T cell infiltration, in diseases such as ovarian cancer (2), date to the early 2000s and suggested an important but unclear role for the immune response in advanced cancer. Subsequent analyses from tumor biopsies of patients with melanoma treated in cancer vaccine clinical trials suggested two broad tumor microenvironmental phenotypes (3). In tumors from patients that appeared to benefit, there was an association with the presence of infiltrating T cells. Early use of RNA micro-arrays further defined these tumor microenvironments as being associated with the presence of type I/II interferon (IFN) associated gene transcripts as well as chemokines related to T cell recruitment (4). This phenotype was described as the T cell-inflamed tumor microenvironment and set the stage as a precursor model to that described by Ayers et al. for pembrolizumab (5) and subsequently used by Ott et al. in KEYNOTE-028. This T cell-inflamed GEP is an 18-gene signature that was derived from cluster analysis of pan-cancer treatment response across several KEYNOTE trials (5), including IFNγ responsive genes related to chemokine expression, antigen presentation, and adaptive immune resistance. Given these characteristics, the association of PD-L1 and the T cell-inflamed GEP should come as little surprise given the known role for IFNγ in regulating the induction of PD-L1 in multiple cell populations. In contrast, non-T cell-inflamed tumors lack IFNγ associated gene expression, with T cells being either absent or very infrequent within or around the tumor. Defects in both T cell priming and migration of T cells may lead to this phenotype (6) and these tumors are not responsive to checkpoint inhibition (1,5).
Concurrent with Ott et al., Cristescu et al. performed an analysis of TMB and T cell-inflamed GEP across four KEYNOTE trials (KEYNOTE-001, -006, -012, -028) where whole exome sequencing or immune gene expression were available (7). T cell-inflamed GEP score had a significant correlation with best overall response, as did TMB. Correlation of TMB and GEP was low or absent across the investigated patient cohorts, reinforcing that TMB and GEP quantify different aspects of tumor biology and clinical response to checkpoint inhibition. Cristescu et al. then analyzed GEPhi/lo in combination with TMBhi/lo, to assess if the biomarkers were more sensitive in combination. GEPhi plus TMBhi had highest response rate while GEPlo plus TMBlo had the lowest. Additionally, other reported mutational signatures such as predicted neoantigens, smoking status, APOBEC-driven mutations, UV light exposure, DNA transversions, homologous recombination deficiency and MSI demonstrated no meaningful improvement over TMB alone (7). Multiple studies have now demonstrated that neither TMB nor neoantigen load correlate with tumor-immune cell infiltration (or T cell-inflamed GEP) (8,9) and the absolute number of predicted or known tumor antigens has been found to be comparable between T cell inflamed and non-T cell-inflamed tumors (10). Taken together, these studies by Ott et al. and others demonstrate the orthogonal relationship between measures of tumor antigenicity and tumor infiltration in predicting response to PD1 immunotherapy across tumor types.
The relationship of TMB and IFNγ GEP is illustrated in Figure 1, with responders to checkpoint inhibition generally having either, or both, antigens and tumor infiltration present, with non-responders demonstrating low antigen burden and/or immune infiltration. This model integrating TMB and IFNγ GEP as primary biomarkers has the potential to improve the design or interpretation of novel immunotherapy clinical trials. For example, in a single arm phase II trial it might be the case that an improved response rate relative to historical controls is observed for the combination of a PD1 antibody with a second agent. If tumors from the responding patients were profiled as GEPhi/TMBhi (quadrant B) at baseline however, it would be difficult to interpret the improved response rate relative to PD1 antibody alone. This would be especially the case for combination approaches involving a second agent without substantive monotherapy activity. This consideration might be seen as timely given the recent failure of ECHO301/KEYNOTE-252, where any incremental benefit from epacadostat had not been established on a translational level (11). If profiling of tumors via TMB and GEP had demonstrated responding patients perhaps from quadrant A to B (GEPhi/TMBlo to GEPhi/TMBhi) or D to B (GEPlo/TMBhi to GEPhi/TMBhi) perhaps a better rationale, beyond response rate from a small number of patients in phase II, would have supported moving to phase III.
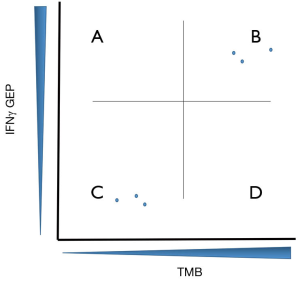
This model of GEP/TMB can also be used to integrate rational selection into clinical trial designs. To date, many immunotherapy drugs advanced in clinical trials of PD1 refractory patients have built on PD1 antibody as a backbone and combined with a second agent, usually an IFN associated gene target. Most of these trials have also pursued patient enrollment in a biological unselected fashion and, rather unsurprisingly, have had unimpressive results (6). An alternative approach might be to place the drug target into the quadrants of Figure 1 and design patient selection criteria around this. Two examples of success supporting this approach can be found in the development of the LAG3 antibody relatlimab and the TIM3 antibody TSR-022. Ascierto et al. have reported an intriguing response rate in patients with melanoma, progressed on PD1, with the combination of nivolumab with relatlimab. These responses are isolated only to those patients with tumors harboring high levels if LAG3 expression by immunohistochemistry (12). Similarly, Davar et al. described a population of responding patients with NSCLC, progressed on prior PD1 antibody but that retained PD-L1 expression, who responded to the combination of the PD1 antibody dostarlimab and TSR-022 (13). As LAG3 and TIM3 are IFN responsive genes, the patient harboring these tumors would have been characterized into quadrants A or B of Figure 1 highlighting the potential utility of this sort of selection mechanism. Profiling patient tumors into quadrant D (GEPlo/TMBlo) identifies the very high need patient population of immunotherapy non-responders and could emphasize a biological population to target with antigen-specific approaches such as cellular therapies or T cell re-directing therapies and perhaps innate immune stimulators such as agonists of Toll-like receptor or STING agonists. Examples to highlight demonstrating conceptual success using this model might be intra-pleural administration of mesothelin specific chimeric antigen receptor T cells in mesothelioma (14) and the bispecific T cell-redirecting molecule tebentafusp in uveal melanoma (15). Both of these tumor types generally exist in quadrant D (GEPlo/TMBlo) (10) of Figure 1 however these immunotherapy approaches appear to engender treatment benefit for patients with these tumors.
Immunotherapy has rapidly come forward as an essential element in the treatment of advanced cancer however a major gap remains surrounding patient selection and biomarker integration into clinical practice. The work by Ott et al. in KEYNOTE-028 lay important groundwork toward the rational integration of PD-L1, TMB and T cell-inflamed GEP in standard treatment as well as next generation immuno-oncology clinical trials. KEYNOTE-028 identifies that many tumor types have the potential to respond to PD1 immunotherapy but that multi-dimensional biomarker analysis will be needed to optimize patient level benefit. Using these biomarkers to predict response is an important goal however one wonders whether the use of these markers to note which patients are very unlikely to respond might be an at least as important a goal. Comprehensive analysis of PD-L1, TMB and T cell-inflamed GEP should be a priority in all immunotherapy trials moving forward. This would facilitate the identification of therapeutic approaches in tumors that are low for these biomarkers and nominate new biomarkers of relevance for tumors demonstrated as biomarker high but that do not respond to anti-PD-1 immunotherapy.
Acknowledgments
Funding: JJ Luke acknowledges Department of Defense Career Development Award (W81XWH-17-1-0265).
Footnote
Provenance and Peer Review: This is an invited article commissioned and reviewed by the Academic Editor Dr. Jiarui Li (Department of Medical Oncology, Peking Union Medical College Hospital, Beijing, China).
Conflicts of Interest: JJ Luke declares Data and Safety Monitoring Board: TTC Oncology; Scientific Advisory Board: 7 Hills, Actym, Alphamab Oncology, Array, BeneVir, Mavu, Tempest; Consultancy: Aduro, Astellas, AstraZeneca, Bayer, Bristol-Myers Squibb, Castle, CheckMate, Compugen, EMD Serono, IDEAYA, Immunocore, Janssen, Jounce, Leap, Merck, Mersana, NewLink, Novartis, RefleXion, Spring Bank, Syndax, Tempest, Vividion, WntRx; Research Support: (all to institution for clinical trials unless noted) AbbVie, Array (Scientific Research Agreement; SRA), Boston Biomedical, Bristol-Myers Squibb, Celldex, CheckMate (SRA), Compugen, Corvus, EMD Serono, Evelo (SRA), Delcath, Five Prime, FLX Bio, Genentech, Immunocore, Incyte, Leap, MedImmune, Macrogenics, Novartis, Pharmacyclics, Palleon (SRA), Merck, Tesaro, Xencor; Travel: Array, AstraZeneca, Bayer, BeneVir, Bristol-Myers Squibb, Castle, CheckMate, EMD Serono, IDEAYA, Immunocore, Janssen, Jounce, Merck, Mersana, NewLink, Novartis, RefleXion; Patents: (both provisional) Serial #15/612,657. M Jameson-Lee has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ott PA, Bang YJ, Piha-Paul SA, et al. T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol 2019;37:318-27. [Crossref] [PubMed]
- Zhang L, Conejo-Garcia JR, Katsaros D, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003;348:203-13. [Crossref] [PubMed]
- Harlin H, Meng Y, Peterson AC, et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res 2009;69:3077-85. [Crossref] [PubMed]
- Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J 2010;16:399-403. [Crossref] [PubMed]
- Ayers M, Lunceford J, Nebozhyn M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930-40. [Crossref] [PubMed]
- Trujillo JA, Sweis RF, Bao R, et al. T Cell-Inflamed versus Non-T Cell-Inflamed Tumors: A Conceptual Framework for Cancer Immunotherapy Drug Development and Combination Therapy Selection. Cancer Immunol Res 2018;6:990-1000. [Crossref] [PubMed]
- Cristescu R, Mogg R, Ayers M, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018. [Crossref] [PubMed]
- Hellmann MD, Nathanson T, Rizvi H, et al. Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 2018;33:843-52.e4. [Crossref] [PubMed]
- Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017;171:934-949.e16. [Crossref] [PubMed]
- Spranger S, Luke JJ, Bao R, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci U S A 2016;113:E7759-68. [Crossref] [PubMed]
- Labadie BW, Bao R, Luke JJ. Reimagining IDO Pathway Inhibition in Cancer Immunotherapy via Downstream Focus on the Tryptophan-Kynurenine-Aryl Hydrocarbon Axis. Clin Cancer Res 2019;25:1462-71. [Crossref] [PubMed]
- Ascierto PA, Melero I, Bhatia S, et al. Initial efficacy of anti-lymphocyte activation gene-3 (anti–LAG-3; BMS-986016) in combination with nivolumab (nivo) in pts with melanoma (MEL) previously treated with anti–PD-1/PD-L1 therapy. J Clin Oncol 2017;35:9520. [Crossref]
- Davar D. Abstract O21: A phase 1 study of TSR-022, an anti-TIM-3 monoclonal antibody, in combination with TSR-042 (anti-PD-1) in patients with colorectal cancer and post-PD-1 NSCLC and melanoma. SITC annual meeting 2018.
- Adusumilli PS, Zauderer MG, Rusch VW, et al. Regional delivery of mesothelin-targeted CAR T cells for pleural cancers: Safety and preliminary efficacy in combination with anti-PD-1 agent. J Clin Oncol 2019;37:2511. [Crossref]
- Damato BE, Dukes J, Goodall H, et al. Tebentafusp: T Cell Redirection for the Treatment of Metastatic Uveal Melanoma. Cancers (Basel) 2019. [Crossref] [PubMed]


