
Role of image-guided biopsy and radiomics in the age of precision medicine
Introduction
The multi-scale character of cancer heterogeneity is a widely described in the literature as a determining parameter for patient survival (1). Heterogeneity or clinical-pathological variability prevents an effective standardized cancer treatment regimen, and necessitates a patient-centered and patient-tailored approach to comprehensive cancer management.
Extensive genetic and phenotypic variation between tumors results in this heterogeneity, and can be observed at different scales (1). Variation can be present between patients (inter-patient) and within the same patient (intra-patient). Tumoral heterogeneity within the same patient may present within the primary tumor (intra-tumoral), between metastatic lesions (inter-metastatic), and even within a single metastasis (intra-metastatic) (2).
It is important to note that intra- and inter-tumor heterogeneity also undergoes temporal variation due to genomic instability and is recognized as an important factor leading to cancer treatment failure and poor prognosis (3,4).
A better understanding of the phenomena underlying tumor heterogeneity is crucial to improve cancer management (5,6). The quantification of heterogeneity relies on identification of various biomarkers, by the use of either tissue biopsy or medical imaging features. Spatial and temporal heterogeneity can be accurately estimated by sophisticated genomic analyses that generally require repeated tumor biopsies. While the image-guided biopsies offer excellent spatial resolution for tissue analysis on a cellular scale and allow proteomic, genetic and molecular sequencing (7); however, biopsy is limited by risks of invasive procedures and focal sampling errors and limitation by tumoral characteristics such as small size, location, or heterogenous necrosis (2,8).
In the search for a non-invasive method to characterize tumor, researchers have turned to medical imaging as a repeatable alternative to quantify tumoral heterogeneity, albeit with a lower spatial resolution. The benefits of non-invasive imaging compared to invasive image-guided biopsy include reduced inherent risk to the patient and the potential to characterize the tumor as a whole as opposed to a focal biopsy. In addition, imaging can assess the temporal course of tumoral heterogeneity along the treatment sequence or disease progression to guide treatment. The limitations of medical imaging include radiation exposure, imaging contrast resolution limitations by imaging modality, and specific imaging modality restrictions such as magnetic resonance imaging (MRI) safety restrictions in patients with implants.
Recent advances in medical imaging informatics have allowed the extraction of advanced quantitative imaging biomarkers without any additional examination or cost. Biomarkers identified on imaging alone can describe the tumoral phenotype and extract metrics that measure in vivo the internal organization and evaluate the spatial spread of the malignant process as well as the surrounding tissue (host). Biomarker extraction from diagnostic images is referred to as radiomics or Image omics. This advanced quantitative technique was first introduced to decode the genomic activity of tumors (9,10) and then applied to a large number of pathologies and different imaging modalities, predominantly computed tomography (CT) and MRI.
The term Radiomics was introduced by Gillies et al. in 2010 (11) and adopted by Lambin et al. in 2012 (12). Radiomics can guide the treatment of oncology patients, by classifying tumors by stage, histology or molecular profile (13,14), or to predict response to treatment (15). This technique has also been proposed to focal treatments, such as radiotherapy (16). Used in parallel or in addition to conventional biomarkers from biopsy and clinical data, radiomics is currently a major research topic for the development of personalized medicine, as all digitized images obtained in medical imaging can benefit from radiomics analysis based on the principle of texture.
The purpose of this article is to describe the current role of tumor biopsies and the principles of radiomics analysis in the era of precision medicine, and to present potential synergies to improve patients’ outcomes.
Biopsies and precision medicine
Percutaneous image-guided tumor biopsy remains the standard method to acquire samples for analysis, of both tumoral and non-tumoral tissue. Percutaneous, image-guided biopsies in many institutions have obviated the need for surgical excisional biopsy. Image-guided biopsy provides a minimally invasive technique with very good results for routine diagnostic purposes, as well as for precision cancer diagnostics. While overall less invasive than surgical biopsy, image-guided needle biopsy still presents potential procedure related risks and complications (17), reported to be below 5–10%, which must be weighed against the potential gains of tissue sampling that include acquisition of predictive biomarkers to guide therapy selection between conventional anticancer treatments, targeted therapies, and immunotherapies.
The pre-biopsy work-up includes careful evaluation of diagnostic imaging for procedural planning. Pre-procedure imaging must be obtained to ensure that the target lesion is well visualized. Image-guided biopsy may be performed with the modality that best visualizes the lesion, and most commonly employs ultrasound or CT guidance, but may also utilize fluoroscopic, MRI or positron emission tomography (PET)-CT guidance. In addition to lesional visualization, a safe trajectory to the lesion must be visible through the surrounding soft tissue structures to minimize complication risks. Lastly, before proceeding with biopsy, evaluation of imaging may identify higher density of lesional tissue in certain portions of the tumor, which may improve viability of sampling. For example, increases nodular density, vascularity, or perfusion in the periphery of a lesion may guide the proceduralist from avoiding sampling centrally necrotic debris. In an ancillary study of the MOSCATO trial, a molecular trial based on on-purpose biopsies, many predictive factors for high tumor cellularity were analyzed comprehensively (18). The organ (other than bone), the number of samples (≥6), and the use of contrast enhanced ultrasound were predictive for higher cellularity, in order to perform comparative genomic hybridization and next-generation sequencing.
Complications of image-guided needle biopsy are reported at approximately 10%; however, the majority of these complications are minor. In evaluation of CT guided complications from percutaneous image-guided needle biopsy of tumors, a recent large prospective study reported grade 3 (CTCAE) complications to occur in only 1.6% of the patients and no grade 4 or 5 complications (8). The only predictive factor of severe complication was the organ biopsied with an odds ratio =27.23 (4.93–242.76), mainly from pneumothorax complications after lung tumor biopsy. Despite this higher risk for lung tumor biopsy, the treatment for the complication is typically placement of a small caliber (8–10 Fr) chest tube for 1–2 nights, a minor inconvenience compared to the routine injury and complications from open surgical lung biopsy. Sequential and multisite biopsies have also been reported with high yield and acceptable low complication risks, specifically in the setting of clinical research protocols performed by experienced operators (19,20).
After acquisition of sample, the next critical aspect is the analysis and sample post-processing. Recent technical developments have improved the outcome of analysis, particularly for needle biopsy samples that provide relatively small number of cells. These laboratory advancements include deep sequencing with single-cell analysis, RNA sequencing, and whole exome sequencing. In addition, patient-derived xenograft (PDX) models (21), can be applied to develop precision medicine.
Lastly, the convenience of needle sampling and immediate local ablative therapy should be highlighted. Biopsies and focal image-guided treatments such as percutaneous ablations can be performed during the same procedural setting, without increasing the puncture-related risks. For example, samples may be obtained and the lesion may be definitely ablated with percutaneous thermal ablation, all via the same coaxial needle access (22,23).
The main drawbacks to the application of percutaneous biopsies for precision medicine, beside the inherent, albeit low, risks to needle guided intervention, remain the inherent procedural logistics, required technical expertise, and the related costs (24).
Radiomics and precision medicine
Radiomics translates images computationally into quantitative complex data. The imaging features that are extracted include tissue intensity, shape, and texture. The detailed description and categorization of these quantitative complex data are not routinely part of the radiological lexica due to the inherent difficulty in discretion by the naked eye. Several computation models seek to improve characterization through dependable algorithms using high dimensional data that allows for more in-depth characterization of the tumor phenotype (25-27). The underlying assumption is that the tumoral imaging patterns reflect not only tissue architecture, but also cellular and molecular composition. Radiomics provides the potential for a “virtual biopsy”. The advantages of radiomics include: (I) non-invasive process, (II) ability to simultaneous provide tissue composition and tumoral spatial heterogeneity by analysis of the lesion patterns and spread over time, and (III) reproducibility of processes to monitor the evolution of the disease, including natural history, temporal heterogeneity, prediction of treatment response and modifications induced by therapy.
Since oncologic imaging is already heavy integrated to routine clinical management, a vast amount of data is already available to facilitate development of the radiomics field at each step of the patient management: diagnosis, staging, and follow up. This high volume of imaging has already demonstrated improvements in cancer care. A National Lung Cancer Screening Trial (NLST) that included 53,000 patients demonstrated that low-dose CTs interpreted by radiologists have shown to be associated with mortality reduction. Unfortunately, there was a high number of false-positive imaging interpretation (28), which might be effectively improved with further application of radiomics that has accuracies of 80% and 79%, at 1 and 2 years respectively, for predicting nodules that will become cancerous (29). With improvements, radiomics will be more widely applied towards precision medicine, through the characterization of the tumor biology (25).
Radiomics also has the capability to analyze both temporal and spatial heterogeneities through quantitative serial data evaluation. For example, in CTs of lung adenocarcinomas, entropies of the tumor core and boundary regions were computed separately, with results showing that a higher ratio between the two are associated with poorer outcome (30). This method evaluated several volumes of interest in one lesion to provide an in-depth characterization that has also been applied in other studies to show characterization of intratumoral and peritumoral features may improve treatment outcome predictions (31,32).
Radiomics may also have potential to characterize additional tumor oncogenic process, also known as the hallmarks of cancer (33,34). Indeed, tumor angiogenesis has been associated with contrast enhancement imaging phenotype in glioblastomas (10), as well with more advanced texture features in lung (35) or breast cancers (36). Hypoxia has also been investigated and shown to be associated with multi-categorical quantitative imaging features in lung cancers (37,38), and similar findings were published regarding features associated with cell proliferation genes (9). Other studies have shown associations between radiomics features and immune infiltration or immune pathways (31,32,39). For example, radiomics may be use to predict CD8 T cell genes expression, which might serve as a prognostic factor for patients treated with immunotherapy (32).
The applications above apply global radiomics methods to assess imaging features from a volume of interest (VOI). Radiomics may also be applied on a per-voxel basis. This “local radiomics” allow the generation and the visualization of novel tumoral maps (15,40,41) similar to functional imaging such as PET-CT, but with the advantage of the resolution of anatomical imaging resolution and the absence of a PET tracer (42). The resultant feature maps may detect more aggressive areas of the tumor, which can help to guide biopsies and also local treatments such as percutaneous thermal ablation or radiotherapy to targeted volumes to the radiomics map (radiomic-target volume) in dose painting strategies (15,43).
While radiomics seems a very promising tool with the ultimate goal to qualify radiomic features as biomarkers for clinical care, further developments and improvements are needed to achieve reliable and clinically applicable results (44,45). Studies have demonstrated that reproducibility is affected by imaging acquisition techniques/parameters, reconstruction algorithms (smooth and sharp), contrast-enhancement quality (46), and segmentation. These findings raise awareness of the importance of properly setting imaging acquisition parameters, especially in the context of radiomics approaches used across multiple institutions. Today the level of evidence of the different studies still remains low and further clinical studies are needed to homogenize imaging protocols and to improve feature reproducibility (47,48). Prospective clinical trials are also needed to validate the findings for different tumor types and subtypes, as well as different imaging modalities. In order to help to improve the quality and the level of evidence of the numerous studies in this field of research, evaluation criteria and reporting guidelines have been published (47).
Potential synergies of image-guided biopsies and radiomics
While both image-guided needle biopsy and radiomics techniques share the same objectives, the approaches are very distinct. There are potential advantages to combining techniques (Figure 1).
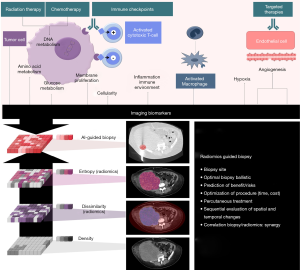
Table 1 summarizes the advantages and limitations of image-guided biopsies and radiomics, and potential synergies to better drive precision medicine.
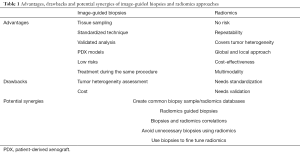
Full table
The most immediately application of radiomics for image-guided biopsies would be to precisely guide biopsy of a specific part of a tumor or peritumoral tissue. “Radiomics-guided” biopsies could improve biopsy yield through identification of increase detection of molecular alterations or resistance mutation. Conversely, biopsy-proven radiomics could provide a means to validate radiomic patterns of interest, and go beyond the statistical correlations with phenotypes or clinical features by matching specific molecular findings. Well-structured databases that correlate radiological findings with pathological samples and clinical outcome required extensive database collection, which could be retrospectively or prospectively gathered in tertiary cancer centers with early phase drug development units.
Another application could be for patients that can undergo a biopsy because of the tumor location or other contraindications. For glioblastomas, for instance, radiomics approaches have also been widely deployed for the diagnosis and treatment plan. Several signatures were identified and associated with methylation, age-related patterns and prognosis factors (49). Although these technologies should be validated prospectively, there is significant evidence to suggest that radiomics approaches could provide virtual biopsy of the tumor heterogeneity and vascularity in glioblastoma patients.
Another example of risk optimization is the radiomics-based clinical decision algorithm applied to cirrhotic patients requiring liver biopsy (European guidelines) or wait-and-see strategy (American guidelines). A signature using a single feature was validated in a multicenter retrospective cohort to diagnose hepatocellular carcinoma (HCC) and therefore improve their management and outcome (50), showing perfectly the incremental value of computational approach extracted from standard of care triphasic CT-scans. While expert visual assessment using current guidelines cannot accurately differentiate HCC from differential diagnoses, this process can be automated. Narrow artificial intelligence proved to excel in specific task such as quantifying a “washout” pattern between arterial and portal venous phases that is a hallmark of this disease.
The analysis of source imaging data from large randomized trials offers a rich and reusable, but largely untapped, resource for future research on novel trial-level response and progression imaging metrics (51). Once validated, imaging biomarkers associated with poor prognosis could be leveraged to detect tumor lesions with aggressive phenotype/high-risk and therefore improve radiomics-guided biopsies. In addition, this database evaluation could be applied to better determine when biopsy is indicated, and which tissue sampling is unnecessary (52). For example, biopsies could be pursued in specific cases when radiomic patterns are not concordant to clinical outcomes or to elucidate new patterns that could be of interest.
Conclusions
Percutaneous image-guided needle biopsies and radiomics are key players in the age of precision medicine for patient-tailored therapy, with specific limitations and advantages to both techniques. Future advances in radiomics hold great promise, but will require extensive database collection to correlate diagnostic imaging characteristics with tissue sampling and clinical outcome. A combined approach of biopsy and radiomics may ultimately provide the optimal tumoral characterization with minimal risks to most appropriately guide specialized therapy.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Allison KH, Sledge GW. Heterogeneity and cancer. Oncology (Williston Park) 2014;28:772-8. [PubMed]
- Dercle L, Ammari S, Bateson M, et al. Limits of radiomic-based entropy as a surrogate of tumor heterogeneity: ROI-area, acquisition protocol and tissue site exert substantial influence. Sci Rep 2017;7:7952. [Crossref] [PubMed]
- Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 2012;12:323-34. [Crossref] [PubMed]
- Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338-45. [Crossref] [PubMed]
- Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin Cancer Res 2015;21:1258-66. [Crossref] [PubMed]
- Robertson-Tessi M, Gillies RJ, Gatenby RA, et al. Impact of metabolic heterogeneity on tumor growth, invasion, and treatment outcomes. Cancer Res 2015;75:1567-79. [Crossref] [PubMed]
- Massard C, Michiels S, Ferte C, et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov 2017;7:586-95. [Crossref] [PubMed]
- Prud'homme C, Deschamps F, Allorant A, et al. Image-guided tumour biopsies in a prospective molecular triage study (MOSCATO-01): what are the real risks? Eur J Cancer 2018;103:108-19. [Crossref] [PubMed]
- Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25:675-80. [Crossref] [PubMed]
- Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105:5213-8. [Crossref] [PubMed]
- Gillies RJ, Anderson AR, Gatenby RA, et al. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol 2010;65:517-21. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Liang C, Huang Y, He L, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I-II and stage III-IV colorectal cancer. Oncotarget 2016;7:31401-12. [Crossref] [PubMed]
- Wu W, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol 2016;6:71. [Crossref] [PubMed]
- Shiradkar R, Podder TK, Algohary A, et al. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol 2016;11:148. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol 2013;31:17-22. [Crossref] [PubMed]
- Tacher V, Le Deley MC, Hollebecque A, et al. Factors associated with success of image-guided tumour biopsies: Results from a prospective molecular triage study (MOSCATO-01). Eur J Cancer 2016;59:79-89. [Crossref] [PubMed]
- Tsai EB, Pomykala K, Ruchalski K, et al. Feasibility and safety of intrathoracic biopsy and repeat biopsy for evaluation of programmed cell death ligand-1 expression for immunotherapy in non-small cell lung cancer. Radiology 2018;287:326-32. [Crossref] [PubMed]
- Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012;18:6373-83. [Crossref] [PubMed]
- Hidalgo M, Amant F, Biankin AV, et al. Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 2014;4:998-1013. [Crossref] [PubMed]
- Tselikas L, de Baere T, Deschamps F, et al. Diagnostic yield of a biopsy performed immediately after lung radiofrequency ablation. Eur Radiol 2017;27:1211-7. [Crossref] [PubMed]
- Schneider T, Puderbach M, Kunz J, et al. Simultaneous computed tomography-guided biopsy and radiofrequency ablation of solitary pulmonary malignancy in high-risk patients. Respiration 2012;84:501-8. [Crossref] [PubMed]
- Pagès A, Foulon S, Zou Z, et al. The cost of molecular-guided therapy in oncology: a prospective cost study alongside the MOSCATO trial. Genet Med 2017;19:683-90. [Crossref] [PubMed]
- Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191-206. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- National Lung Screening Trial Research Team, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Hawkins S, Wang H, Liu Y, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol 2016;11:2120-8. [Crossref] [PubMed]
- Grove O, Berglund AE, Schabath MB, et al. Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. PLoS One 2015;10:e0118261. [Crossref] [PubMed]
- Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res 2017;19:57. [Crossref] [PubMed]
- Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Ganeshan B, Abaleke S, Young RC, et al. Texture analysis of non-small cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging 2010;10:137-43. [Crossref] [PubMed]
- Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016. [Crossref] [PubMed]
- Ganeshan B, Goh V, Mandeville HC, et al. Non-small cell lung cancer: histopathologic correlates for texture parameters at CT. Radiology 2013;266:326-36. [Crossref] [PubMed]
- Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology 2012;264:387-96. [Crossref] [PubMed]
- Grossmann P, Stringfield O, El-Hachem N, et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017. [Crossref] [PubMed]
- Larroza A, Moratal D, Paredes-Sanchez A, et al. Support vector machine classification of brain metastasis and radiation necrosis based on texture analysis in MRI. J Magn Reson Imaging 2015;42:1362-8. [Crossref] [PubMed]
- Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of local anisotropic gradient orientations (CoLlAGe): a new radiomics descriptor. Sci Rep 2016;6:37241. [Crossref] [PubMed]
- Reuzé S, Schernberg A, Orlhac F, et al. Radiomics in nuclear medicine applied to radiation therapy: methods, pitfalls, and challenges. Int J Radiat Oncol Biol Phys 2018;102:1117-42. [Crossref] [PubMed]
- Sun R, Orlhac F, Robert C, et al. In regard to Mattonen et al. Int J Radiat Oncol Biol Phys 2016;95:1544-5. [Crossref] [PubMed]
- Huang Q, Lu L, Dercle L, et al. Interobserver variability in tumor contouring affects the use of radiomics to predict mutational status. J Med Imaging (Bellingham) 2018;5:011005. [PubMed]
- Zhao B, Tan Y, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 2016;6:23428. [Crossref] [PubMed]
- Ma J, Dercle L, Lichtenstein P, et al. Automated identification of optimal portal venous phase timing with convolutional neural networks. Acad Radiol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- Park JE, Kim D, Kim HS, et al. Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol 2020;30:523-36. [Crossref] [PubMed]
- Sinigaglia M, Assi T, Besson FL, et al. Imaging-guided precision medicine in glioblastoma patients treated with immune checkpoint modulators: research trend and future directions in the field of imaging biomarkers and artificial intelligence. EJNMMI Res 2019;9:78. [Crossref] [PubMed]
- Mokrane FZ, Lu L, Vavasseur A, et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur Radiol 2020;30:558-70. [Crossref] [PubMed]
- Dercle L, Connors DE, Tang Y, et al. Vol-PACT: a foundation for the NIH public-private partnership that supports sharing of clinical trial data for the development of improved imaging biomarkers in oncology. JCO Clin Cancer Inform 2018;2:1-12. [Crossref] [PubMed]
- Hu T, Wang S, Huang L, et al. A clinical-radiomics nomogram for the preoperative prediction of lung metastasis in colorectal cancer patients with indeterminate pulmonary nodules. Eur Radiol 2019;29:439-49. [Crossref] [PubMed]


