
Pancreatic adenocarcinomas without KRAS, TP53, CDKN2A and SMAD4 mutations and CDKN2A/CDKN2B copy number alterations: a review of the genomic landscape to unveil therapeutic avenues
Highlight box
Key findings
• Pancreatic cancers without the common alterations in KRAS, TP53, CDKN2A and SMAD4 represent a genomically stable minority of cases, some having alterations in other oncogenes and tumor suppressors of pathogenic interest.
What is known and what is new?
• Although pancreatic adenocarcinoma is a cancer with few molecular alterations, recurrent mutations in oncogenes and tumor suppressors are present. We report here that some of the pancreatic cancers that present without any of the common molecular alterations, still possess other mutations such as in DNA Damage Response genes or in epigenetic modifiers.
What is the implication and what should change now?
• Opportunities for treatment of pancreatic cancers without the common molecular alterations of the disease may include targeting the DNA damage response and the epigenetic machinery with drugs already being used or in development.
Introduction
Pancreatic cancer is a gastrointestinal malignancy with rising incidence and with a 5-year survival rate of less than 10% (1). The median survival of metastatic disease is about 6 months. Prevalence is highest in the developed world compared with low-income countries and is only slightly higher in men compared to women (2). In 2019, pancreatic adenocarcinoma accounted for 45,750 deaths in the United States of America (1). Despite representing only the thirteenth most prevalent cancer worldwide, accounting for 458,000 cases, pancreatic cancer is the seventh cancer in the mortality list (3). Pancreatic cancers usually present in advanced stage due to early dissemination and lack of symptoms when in earlier stages. Moreover, despite improvements in chemotherapy treatments and management in recent years with the introduction of more effective chemotherapy regimens, resistance to radiotherapy and chemotherapy contributes to increased mortality (4-7). Thus, there is a pressing need for improved systemic therapies based on the clarification of pancreatic molecular carcinogenesis that has been the result of genomic cancer research over the last decades (8).
The molecular pathology of pancreatic adenocarcinomas is characterized by few prevalent mutations associated with a low overall tumor mutation burden (TMB). Only 4.6% of pancreatic adenocarcinomas of TCGA cohort have a TMB above 80 (9). Few cancer-associated genes display recurrent mutations in pancreatic cancers. Oncogene KRAS mutations are the most prevalent, encountered in up to 90% of pancreatic cancers, followed by mutations in tumor suppressor TP53, encountered in up to three fourths of pancreatic cancers. Two additional tumor suppressor genes, CDKN2A, encoding for cell cycle inhibitor p16 and the gene encoding for transforming growth factor beta (TGFβ) pathway signal transducer SMAD4, are mutated in about 20% of cases in pancreatic cancers (9,10). Copy number alterations are also less frequent in pancreatic cancers compared with other carcinomas. The most frequently copy altered locus, showing deep deletions, is at chromosome 9p21.3, where CDKN2A is located. The same locus also encodes for the p53 positive regulator p14ARF, which is transcribed by the same sequence with an alternative reading frame. The locus is deleted in 10% to 25% of pancreatic cancer cases. Besides these five recurrent gene alterations, no other oncogene or tumor suppressor is commonly altered in pancreatic cancers. A minority of pancreatic cancers harbor no molecular alterations in any of these genes. The current investigation examines the molecular abnormalities present in these cases characterized by absence of typical pancreatic cancer alterations, for discovery of less common lesions present in these cases that may be associated with an alternative cancer pathogenesis and may serve as targets for novel therapies. Cases of pancreatic cancer with absence of the common pathogenic mutations may be inherently resistant to therapies targeting these mutations, should such treatments be developed, and will require alternative therapies based on putative alternative molecular targets.
Methods
Four publicly available genomic series of pancreatic adenocarcinoma patients, published from The Cancer Genome Atlas (TCGA), the MSK-IMPACT study, the pancreatic cancer sub-cohort of a pan-cancer study from China, and the pancreatic cancer cohort from the American Association for Cancer Research (AACR) project GENIE (public v.12), were interrogated for identification and characterization of patients without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B (9,11-13). Analysis of the series of interest were performed in cBioportal for cancer genomics, a genomic studies platform that is freely accessible and allows for interrogation of genomic alterations in any gene of interest, included in the original studies (14). TCGA employs a whole exome sequencing platform, while the three other studies are based on targeted next generation sequencing (NGS) genomic panels (11-13). TCGA uses the Picard pipeline, which stores data of every sequenced sample in BAM (Binary Alignment and Map) files (9). Significantly mutated genes were identified using the MutSig algorithm, version MutSig2CV (15). Analysis of copy number alterations (CNAs) in TCGA is performed with the GISTIC (Genomic Identification of Significant Targets in Cancer) algorithm, in which a score of 2 or above denotes putative amplification of a gene (16). Chromosomal instability of each sample is measured by the Aneuploidy Score (AS), defined as the sum of the number of chromosome arms in each patient sample that displays copy number alterations (gains or losses) (17). A chromosome arm is considered copy number altered based on the length of alterations, as calculated by the ABSOLUTE algorithm from Affymetrix 6.0 SNP arrays (18). The definition of a somatic copy number alteration was set at more than 80% of the length of the arm. Alterations in 20% to 80% of a given arm length were considered inadequate to call, while chromosomal arms with somatic copy number alterations in less than 20% of the arm length were considered not altered. Another measure of chromosomal instability, the Fraction Genome Altered (FGA) was defined as the fraction of log2 copy number gain or loss (>0.2) divided by the total length of the profiled genome. TCGA also includes mRNA expression analysis. The RSEM algorithm was used for normalization of mRNA expression (19).
The MSK-IMPACT study used a targeted hybridization capture-based NGS platform that included 341 genes and in a more updated version, a total 410 genes were included (7). The platform detects all mutations in protein coding regions of these genes, CNAs, selected mutations in promoter regions and selected structural rearrangements (20). The assay targeted 4,976 exons with a mean coverage of 700 [standard deviation (SD): 182]. Single nucleotide variants and insertions/deletions were called using the MuTect and SomaticIndelDetector tools in the Genome Analysis ToolKit (GATK) programming framework (21).
The pan-cancer study from China from which the Chinese pancreatic cancer group was derived, used a targeted NGS panel of 579 cancer-associated genes from Origimed corporation, Shangai, China (12). Variation calling was performed with methods similar to the ones used in the MSK-IMPACT study.
AACR project GENIE included data from eight contributing institutions (5 from USA, 1 institution each from Canada, France and the Netherlands) and the targeted NGS panels used in each institution had a variable number of genes covered (13).
The functional implications of concomitant mutations in included samples were evaluated with the help of OncoKB (22). OncoKB knowledgebase is a database of cancer-related genes and classifies these genes as oncogenes or tumor suppressors.
Statistical analysis
Statistical comparisons of categorical data were carried out using Fisher’s exact test or the χ2 test and comparisons of continuous parameters were compared with the t-test. The Mann-Whitney U test was used to compare median values. All statistical comparisons were considered significant if P<0.05.
Results
Prevalence of common pancreatic adenocarcinoma molecular alterations and of cases without common alterations
Mutations in oncogene KRAS and tumor suppressor TP53 are the most frequent molecular alterations in pancreatic adenocarcinomas, followed by mutations in SMAD4 and CDKN2A genes and deletions of the 9p21.3 locus. Three extensive pancreatic cancer genomic series using targeted gene panels (Pancreatic cohort of the project GENIE, pancreatic cohort of the MSK-IMPACT series and pancreatic cohort of the Chinese Origimed study) show a consistent prevalence of KRAS mutations between 86.6% and 90.6% (Figure 1). The pancreatic study from TCGA, employing a whole exome platform, shows a lower mutation rate at 65.4%. Mutations in TP53 have also more consistent frequency in the three former series with prevalence between 69.5% and 75.5%, while TCGA shows again a lower prevalence at 59.8%. The four studies display a more homogeneous frequency of SMAD4 and CDKN2A mutations around 20% and between 15% and 20%, respectively. The prevalence of deletions of the CDKN2A/CDKN2B locus varies between the series and is the lowest in the Chinese series at 3.5% and the highest in TCGA at 28% (Figure 1). The prevalence of cases without any of the above mutations or deletions is similar in the 3 series, excepting TCGA (5.4% in the Chinese series, 5% in MSK-IMPACT and 8.3% in project GENIE). TCGA, due to lower prevalence of KRAS and TP53 mutations displays a higher rate of cases without any of the common pancreatic cancer alterations at 22.8%.
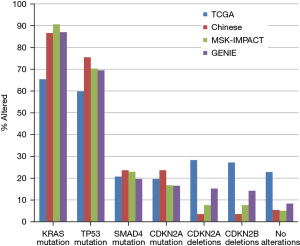
Characteristics and molecular lesions of pancreatic cancers without the common molecular alterations
The clinical and molecular characteristics of pancreatic cancer cases without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B were evaluated in the four cohorts. The 42 patients without these alterations in the TCGA cohort were younger (61.9% younger than 65 years-old versus 52.2% younger than 65 years-old in the whole pancreatic cohort), but this difference was not statistically significant (Fisher’s exact test P=0.3). Similarly, neither the mean age nor the percentage of patients younger than 65 years-old in the Chinese series or the GENIE cohort disclosed any differences between the 2 groups (Table 1). Early (stage I) patients constituted 25% of cases in the group without common alterations in TCGA (versus 11.5% in the whole cohort, Fisher’s exact test P=0.04) (Table 1). In contrast, no differences in the prevalence of localized versus metastatic disease were observed between the 2 groups in the MSK-IMPACT series and in the Chinese cohort (Fisher’s exact test P=0.33 and 0.37, respectively). Genes with mutations in the 42 patients of TCGA cohort encountered in more than one case included ATM and GNAS in 3 cases each (8.1%) (Table 2) and DROSHA, RNF213, KAT6A and LRRK2 in 2 cases each (5.4%). CNAs (all amplifications) in the 42 cases encountered in more than one sample were at genes in loci 12p13.33 (RAD52 and CCND2), 18q11.2, 8p11.23 and 9p13.3, all observed in 2 samples. Besides the cases with ATM mutations no other mutations in DNA damage response (DDR) genes, such as BRCA1, BRCA2, PLB2, BRIP1, or RAD51 homologues were present in the 42 cases. The only CNA in DDR-associated genes was an amplification in BRIP1 observed in a single case. In addition, no alterations in mismatch repair (MMR) genes MSH2, MSH6, MLH1 and PMS2 or in the proofreading polymerases POLE and POLD1 were observed (Table 3). The mean mutation count in the 33 cases of the 42 with available data was 19.6 and no cases had more than 115 mutations (Table 4). Twenty-five of the 33 cases (75.8%) had a low AS of 4 or less.
Table 1
| Characteristic | TCGA | MSK-IMPACT | Chinese Origene | Project GENIE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (n=184) | Without alterations (n=42) | All (n=384) | Without alterations (n=19) | All (n=461) | Without alterations (n=25) | All (n=5,423) | Without alterations (n=448) | ||||
| Age mean ± SD, years | 64.8±11 | 64.5±9.9 | NA | NA | 60.1±9.8 | 59.3±11 | 63.9±10.5 | 61.3±13.7 | |||
| Age (years), n (%) | |||||||||||
| ≤65 | 96 (52.2) | 26 (61.9) | NA | NA | 323 (70.1) | 17 (68.0) | 2,260 (53.2) | 220 (57.1) | |||
| >65 | 88 (47.8) | 16 (38.1) | – | – | 138 (29.9) | 8 (32.0) | 1,988 (46.8) | 165 (42.9) | |||
| NA | – | – | – | – | – | – | 1,175 | 63 | |||
| Sex, n (%) | |||||||||||
| Male | 101 (54.9) | 22 (52.4) | 205 (53.5) | 13 (68.4) | 278 (60.3) | 16 (64.0) | 2,792 (53.2) | 239 (54.2) | |||
| Female | 83 (45.1) | 20 (47.6) | 178 (46.5) | 6 (31.6) | 183 (39.7) | 9 (36.0) | 2,457 (46.8) | 202 (45.8) | |||
| NA | – | 2 | – | – | – | – | 13 | 7 | |||
| Stage, n (%) | |||||||||||
| I | 21 (11.4) | 10 (25.0) | 231 (60.3) | 9 (stages I–III) (47.4) | 100 (23.5) | 6 (27.3) | NA | NA | |||
| II | 151 (82.1) | 29 (72.5) | – | – | 119 (28.0) | 10 (45.4) | – | – | |||
| III | 5 (2.7) | 1 (2.5) | – | – | 43 (10.1) | 0 | – | – | |||
| IV | 5 (2.7) | 0 | 152 (39.7) | 10 (52.6) | 163 (38.4) | 6 (27.3) | – | – | |||
| NA | 2 | 2 | – | – | 36 | 3 | – | – | |||
Numbers in parentheses are percentages and are rounded to the first decimal digit. NA, not available; TCGA, The Cancer Genome Atlas; MSK-IMPACT, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; GENIE, Genomic Evidence Neoplasia Information Exchange.
Table 2
| Mutation | TCGA | MSK-IMPACT | Chinese Origene | Project GENIE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (n=184), n (%) | Without alterations (n=42), n (%) | All (n=384), n (%) | Without alterations (n=19), n (%) | All (n=461), n (%) | Without alterations (n=25), n (%) | All (n=5,423), n (%) | Without alterations (n=448), n (%) | ||||
| ARID1A | 9 (4.9) | 1 (2.7) | 36 (9.4) | 0 | 55 (11.9) | 0 | 427 (8.9) | 23 (6.3) | |||
| GNAS | 7 (3.9) | 3 (8.1) | 13 (3.4) | 3 (15.8) | 4 (0.9) | 0 | 142 (2.6) | 19 (4.3) | |||
| ATM | 8 (4.5) | 3 (8.1) | 11 (2.9) | 0 | 21 (4.6) | 3 (12.0) | 227 (4.4) | 12 (2.8) | |||
| PIK3CA | 5 (2.8) | 0 | 13 (3.4) | 0 | 12 (2.6) | 0 | 163 (3.0) | 12 (2.7) | |||
| BRCA2 | 2 (1.1) | 0 | 13 (3.4) | 0 | 8 (1.7) | 0 | 212 (4.3) | 11 (2.9) | |||
| RNF43 | 11 (6.1) | 0 | 27 (7.0) | 1 (5.3) | 30 (6.5) | 1 (4.0) | 297 (6.3) | 6 (1.7) | |||
| KMT2D | 7 (3.9) | 1 (2.7) | 15 (3.9) | 1 (5.3) | 21 (4.6) | 0 | 291 (6.3) | 6 (1.8) | |||
| TGFBR2 | 8 (4.5) | 1 (2.7) | 13 (3.4) | 1 (5.3) | 22 (4.8) | 0 | 127 (3.6) | 5 (1.1) | |||
| KDM6A | 7 (3.9) | 0 | 13 (3.4) | 0 | 18 (3.9) | 1 (4.0) | 181 (3.9) | 3 (0.9) | |||
| KMT2C | 7 (3.9) | 0 | 12 (3.1) | 1 (5.3) | 12 (2.6) | 0 | 123 (3.7) | 6 (2.7) | |||
Numbers in parentheses are percentages and are rounded to the first decimal digit. TCGA, The Cancer Genome Atlas; MSK-IMPACT, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; GENIE, Genomic Evidence Neoplasia Information Exchange.
Table 3
| Alterations | TCGA | MSK-IMPACT | Chinese Origene | Project GENIE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (n=184), n (%) | Without alterations (n=42), n (%) | All (n=384), n (%) | Without alterations (n=19), n (%) | All (n=461), n (%) | Without alterations (n=25), n (%) | All (n=5,423), n (%) | Without alterations (n=448), n (%) | ||||
| MSH2 mutations | 1 (0.6) | 0 | 1 (0.3) | 0 | 2 (0.4) | 0 | 43 (0.9) | 1 (0.3) | |||
| MSH6 mutations | 1 (0.6) | 0 | 4 (1.0) | 0 | 3 (0.7) | 0 | 69 (1.4) | 2 (0.5) | |||
| MLH1 mutations | 1 (0.6) | 0 | 3 (0.8) | 0 | 5 (1.1) | 0 | 44 (0.8) | 3 (0.7) | |||
| PMS2 mutations | 1 (0.6) | 0 | 0 | 0 | 4 (0.9) | 0 | 28 (0.6) | 0 | |||
| POLE mutations | 2 (1.1) | 0 | 2 (0.5) | 0 | 3 (0.7) | 0 | 70 (1.5) | 2 (0.6) | |||
| POLD1 mutations | 1 (0.6) | 0 | 2 (0.5) | 0 | 3 (0.7) | 0 | 55 (1.3) | 2 (0.7) | |||
| BRAF mutations | 2 (1.1) | 0 | 3 (0.8) | 0 | 13 (2.8) | 0 | 83 (1.5) | 2 (0.5) | |||
| BRAF fusion | 1 (0.5) | 1 (2.4) | 5 (1.3) | 1 (5.3) | NA | NA | 14 (0.3) | 16 (3.5) | |||
| ERBB2 mutations | 2 (1.1) | 0 | 3 (0.8) | 0 | 3 (0.7) | 0 | 68 (1.3) | 6 (1.5) | |||
| ERBB2 amplifications | 9 (4.9) | 0 | 6 (1.6) | 0 | 4 (0.9) | 0 | 41 (0.7) | 2 (0.5) | |||
| FGFR2 mutations | 0 | 0 | 2 (0.5) | 0 | 1 (0.2) | 0 | 28 (0.5) | 1 (0.3) | |||
| FGFR2 amplifications | 1 (0.5) | 0 | 0 | 0 | 0 | 0 | 4 (0.07) | 0 | |||
| FGFR1 amplifications | 7 (3.8) | 2 (4.8) | 6 (1.6) | 0 | 5 (1.1) | 0 | 57 (1.4) | 0 | |||
| FGFR2 fusions | 0 | 0 | 2 (0.5) | 1 (5.3) | NA | NA | 9 (0.2) | 5 (1.2) | |||
| FGFR3 fusions | 0 | 0 | 0 | 0 | NA | NA | 3 (0.1) | 0 | |||
| ALK fusions | 0 | 0 | 1 (0.3) | 0 | NA | NA | 9 (0.2) | 1 (0.3) | |||
| RET fusions | 0 | 0 | 0 | 0 | NA | NA | 1 (0.02) | 0 | |||
| MET fusions | 0 | 0 | 0 | 0 | NA | NA | 3 (0.1) | 1 (0.3) | |||
| ERBB4 fusions | 0 | 0 | 0 | 0 | NA | NA | 1 (0.02) | 0 | |||
| NRG1 fusions | 2 (1.1) | 1 (2.4) | NA | NA | NA | NA | 11 (0.8) | 5 (4.4) | |||
| NTRK1 fusions | 1 (0.5) | 1 (2.4) | 0 | 0 | NA | NA | 6 (0.1) | 1 (0.3) | |||
| NTRK2 fusions | 0 | 0 | 0 | 0 | NA | NA | 2 (0.04) | 0 | |||
| NTRK3 fusions | 1 (0.5) | 0 | 2 (0.5) | 2 (10.5) | NA | NA | 6 (0.1) | 3 (0.9) | |||
Numbers in parentheses are percentages and are rounded to the first decimal digit. NA, not available; TCGA, The Cancer Genome Atlas; MSK-IMPACT, Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets; GENIE, Genomic Evidence Neoplasia Information Exchange.
Table 4
| Characteristic | TCGA | MSK-IMPACT | Chinese Origene | Project GENIE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| All (n=184), n (%) | Without alterations (n=42), n (%) | All (n=384), n (%) | Without alterations (n=19), n (%) | All (n=461), n (%) | Without alterations (n=25), n (%) | All (n=5,423), n (%) | Without alterations (n=448), n (%) | ||||
| TMB | |||||||||||
| Low | 171 (98.3) | 32 (97.0) | 249 (64.8) | 16 (84.2) | 229 (51.1) | 12 (100.0) | 2,736 (53.0) | 138 (73.8) | |||
| Intermediate | 2 (1.1) | 1 (3.0) | 129 (33.6) | 3 (15.8) | 193 (43.1) | 0 | 1,999 (38.8) | 36 (19.3) | |||
| High | 1 (0.6) | 0 | 6 (1.6) | 0 | 26 (5.8) | 0 | 425 (8.2) | 13 (6.9) | |||
| NA | 10 | 9 | 13 | 13 | 263 | 261 | |||||
| FGA | |||||||||||
| <0.08 | 89 (48.6) | 34 (82.9) | 242 (63.0) | 12 (63.2) | NA | NA | 1,588 (57.2) | 121 (75.2) | |||
| 0.08–0.35 | 77 (42.1) | 3 (7.3) | 111 (28.9) | 4 (21.0) | 882 (31.8) | 29 (18.0) | |||||
| >0.35 | 17 (9.3) | 4 (9.8) | 31 (8.1) | 3 (15.8) | 304 (11.0) | 11 (6.8) | |||||
| NA | 1 | 1 | 2,469 | 287 | |||||||
TMB low: <100 for TCGA and ≤5 for others; TMB intermediate: 100–200 for TCGA and 5–10 for others; TMB high: >200 for TCGA and >10 for others. Numbers in parentheses are percentages and are rounded to the first decimal digit. NA, not available; TMB, tumor mutation burden; FGA, Fraction Genome Altered; TCGA, The Cancer Genome Atlas; MSK-IMPACT, Memorial Sloan Kettering- Integrated Mutation Profiling of Actionable Cancer Targets; GENIE, Genomic Evidence Neoplasia Information Exchange.
Pancreatic cancers without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B from the MSK-IMPACT cohort (n=19) showed also mutations in GNAS gene in 3 cases (15.8%) and 2 cases (10.5%) had mutations in BCOR. Several other cancer associated genes assayed in the targeted panel employed in this study, including TGFBR2, BAP1, AXIN1, MDM2, RNF43, NF1, BARD1, PARP1, NFE2L2 and ARAF, had mutations in one case each. The only CNA seen in more than one case was an amplification at locus 12p13, harboring genes RAD52 and CCND2 observed in 2 cases (10.5%). No cases with mutations in DDR-associated genes such as BRCA1, BRCA2, PLB2, BRIP1, and RAD51 or mutations in MMR associated genes MSH2, MSH6, MLH1 and PMS2 and the proofreading polymerases POLE and POLD1 were present among the 19 cancers without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations (Table 2). All 19 cases had low mutation counts of 7 or less and 14 of 19 cases had a low FGA below 0.1 (Table 4).
In the Chinese Origene series, which also employed a targeted genomic panel, 25 patients had pancreatic cancers without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B. Three genes had mutations in more than one case, including mutations in ATM in 3 cases (12%), in BCOR in 2 cases (8%) and in ITK, also in 2 cases. Among DDR associated genes one patient (4%) had a BRIP1 mutation. One case each had a mutation and amplification of LZTR1 gene, encoding for a component of an E3 ligase complex involved in KRAS degradation. All patients with without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B and data available had a low TMB (Table 2). This series provided data on the purity of the samples analyzed. The purity of tumor specimens without common pancreatic gene alterations (mean 34%, Standard deviation: 17.07%) was not different from the purity of the specimens in the entire cohort (mean 35.56%, Standard deviation: 16.75%, t-test P=0.6).
The pancreatic cohort of project GENIE includes 448 patients without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B. Most common mutations present in these cases, observed in more than 2% of patients include mutations in ARID1A (6.3%), GNAS (4.3%), BRAF (3.6%), PRBM1 (3.1%), BRCA2 (2.9%), ATM (2.8%), PIK3CA (2.7%), BCOR (2.5%) and NF1 (2.2%). Overall, none of the genes mutated with a prevalence of 3% or higher in at least 2 of the 4 series examined was significantly more frequently mutated in cases without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B compared with the corresponding whole series (Table 2). Table 3 shows molecular alterations of interest that are potentially enriched in pancreatic cancers without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B and for which there exist targeted therapies. Alterations including fusions involving NRG1, encoding for the EGFR family ligand Neuregulin 1 and fusions involving one of the neurotrophic receptor tyrosine kinase family members, NTRK1, NTRK2 and NTRK3 are observed in about 5% cases.
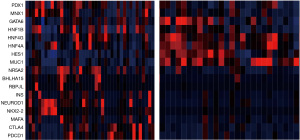
Transcription factors of acinar cell differentiation such as NR5A2, BHLHA15 and RBPJL, together with genes associated with endocrine pancreatic differentiation, such as INS, NEUROD1, NKX2-2, and MAFA were up-regulated in cases without alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B compared with counterparts bearing alterations in one or more of these genes (Figure 2). In addition some cases without alterations present up-regulation of immune checkpoint genes CTLA4 and PDCD1, encoding for PD-1.
Prognosis of pancreatic cancers without the common molecular alterations
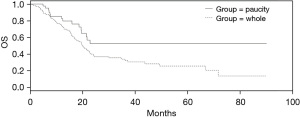
Survival data were available in 2 of the 4 series analyzed, the one from TCGA and the MSK-IMPACT cohort. The overall survival (OS) of patients without the common pancreatic cancer molecular alterations was better than the OS of the whole TCGA pancreatic cancer cohort (Log Rank P=0.03, Figure 3). In contrast, no OS difference between the patients without common alterations and the entire pancreatic group was present in the pancreatic cancer cohort of MSK-IMPACT which had a lower percentage of patients without alterations (Log Rank P=0.45, not shown).
Discussion
Mutations in oncogene KRAS are present in most pancreatic adenocarcinomas and are considered hallmarks of the disease (23). Pancreatic cancer has the highest prevalence of mutations in KRAS among cancers with such mutations, which include colorectal and lung carcinomas. Most commonly KRAS mutations in pancreatic cancer occur at codon 12, with a minority of about 6.5% and 1.2% respectively, occurring at codons 61 and 13 (23). However, these mutations alone may not be sufficient for development of the disease and additional alterations are required for transformation (24). In a mouse model, KRAS activation alone leads only to pancreatic carcinoma in situ, whereas addition of mutant p53 activation leads to early invasive pancreatic carcinogenesis (25). Mutations in TP53 gene encoding for p53, promote pancreatic carcinogenesis through neutralization of the apoptotic and metastasis suppressive functions of p53 (26). Besides KRAS and TP53 mutations, two other commonly mutated genes in pancreatic adenocarcinomas, CDKN2A and SMAD4, lead to deregulation of the cell cycle and the activation of the TGFβ/SMAD pathway, respectively (27). Additionally, the locus of CDKN2A at chromosome 9p is altered by deep deletions in pancreatic cancer, which also affect the overlapping CDKN2B gene, encoding for the p53 positive regulator p14ARF. Among the common molecular alterations of pancreatic cancer, only the KRAS G12C mutations, present in 1% to 2% of cases are currently targeted with available kinase inhibitors. A phase 1/2 trial has shown that inhibitor sotorasib produced a confirmed objective response in 21% of pretreated metastatic pancreatic cancer patients with the specific KRAS mutation (28). Inhibitors for the more common KRAS mutation G12D are also in development (29). Other molecular alterations occur sporadically in pancreatic cancer, with a prevalence in general of less than 8%. The current review of the landscape of pancreatic cancers without the common alterations in KRAS, TP53, SMAD4, CDKN2A and CDKN2B examines whether any of the less frequent molecular alterations is prevalent in these cases. Such alterations could dominantly contribute to the molecular pathogenesis, substituting for the common alterations in these cases. Four extensive pancreatic cancer genomic series formed the basis of this evaluation. Three of the series display similar prevalence of pancreatic cancers without common alterations between 5% and 8.3%, while TCGA shows higher prevalence of such cases (22.8%). This discrepancy is most probably due to the different sequencing platforms used. Targeted gene panels used in the three former studies allow for deeper coverage of the included genes while the whole exome sequencing platform of TCGA employs a lower coverage of each gene and could thus miss altered cases. Another recently published report of pancreatic cancer patients who had genomic analysis with a targeted NGS panel from Caris Life Sciences (Phoenix, AZ) showed that 5.6% of 2,483 patients were KRAS and TP53 wild type (30).
Mutated genes with an overall prevalence between 5% and 10% in pancreatic series (for example ATM, BRCA2, GNAS, and ARID1A) are present in cases without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations. ATM is a pancreatic cancer predisposition gene, with individuals carrying germline pathogenic mutations in the gene having over 6 times the risk of non-carriers to develop pancreatic cancer (31). Results from the four series examined here show that a small number of cases without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations possess ATM mutations. Thus, targeting these mutations therapeutically remains an option in this subset of pancreatic cancers should effective therapies become available. Preclinical studies suggest that although cancer cells with ATM mutations are resistant to PARP inhibitors induced apoptosis, undergoing cell cycle arrest, they are sensitive to a combination of PARP inhibitors with ATR inhibitors (32). The combination of olaparib with ATR inhibitor ceralasertib (AZD6738) led to cell death of ATM deficient cells within 1 to 2 cell divisions, while olaparib monotherapy required multiple replications (33). Besides ATR, another related kinase with a major role in DNA double strand break repair is DNA-PK, which, when recruited in break sites promotes an error prone repair through non-homologous end joining (34). In pancreatic cancer cells and human xenografts with ATM mutations, combinations of PARP inhibitors with ATR and DNA-PK inhibitors were effective in inhibiting growth and in preventing selection of chemotherapy resistant clones (35,36).
The other comparatively frequently mutated DDR associated gene in pancreatic cancer, BRCA2 showed no mutations in pancreatic cancers without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations in 3 of the 4 examined series, but in the project GENIE pancreatic cohort who had higher numbers, 2.9% of cases without such alterations had BRCA2 mutations. This suggests that BRCA2 mutations must be excluded in these cases similar to the rest of pancreatic cancers, in view of the therapeutic option for treatment with PARP inhibitors and the sensitivity to platinum based chemotherapy (37-39).
GNAS encodes for the Gαs sub-unit of a heterotrimeric G protein which signals through the cAMP/protein kinase A (PKA) pathway and the Wnt/beta-catenin pathway (40). GNAS has been identified in the original TCGA report as one of the oncogenes often encountered in pancreatic cancers with wild type KRAS (5). GNAS mutations at codon R201 are common in intraductal papillary mucinous neoplasms (IPMN) of the pancreas, especially the intestinal and colloid subtypes (41). These neoplasms may also possess KRAS mutations, which are not mutually exclusive with GNAS mutations. IPMN derived invasive carcinomas [also called intraductal papillary mucinous carcinomas (IPMC)] possess also GNAS mutations, and are concordant with the precursor lesions for GNAS and KRAS mutations, while accumulating further mutations in TP53, SMAD4 and CDKN2A (42,43). The intestinal and colloid subtypes of IPMC that possess more frequently GNAS mutations have a better prognosis than the usual pancreatic adenocarcinomas which harbor GNAS mutations rarely (44). In contrast the tubular subtype of IPMC display a lower prevalence of GNAS mutations and a higher prevalence of KRAS mutations and behave similarly to pancreatic adenocarcinomas of the usual histology (45). The GNAS/PKA pathway inhibits NOTCH signaling and may decrease cancer cell invasion and metastasis, which is consistent with a better prognosis of cancers with GNAS mutations (46). From a therapeutic point of view, the obvious implication is that, if inhibitors of the pathway become available, their development in established invasive cancers, which possess additional alterations, should proceed with caution. A more straightforward therapeutic candidate for pancreatic cancers with GNAS mutations and colloid histology is immunotherapy with checkpoint inhibitors, as these cancers may be more commonly deficient in MMR or MSI-H than pancreatic cancers with usual histology (47). However, in the pancreatic cancer cases without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations from the four series examined here, most have a low TMB and mutations in MMR associated genes or proofreading polymerases were rare.
ARID1A (AT-rich Interaction Domain-containing protein 1A), encoding for a subunit of chromatin modifier complex SWI/SNF, is mutated in 5% to 10% of pancreatic adenocarcinomas and although only mutated in 1 case (2.7%) without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations from the TCGA cohort it displays a prevalence of 6.3% in the same group of the project GENIE cohort. ARID1A functions to maintain differentiation of pancreatic ductal cells through expression of transcription factor SOX9 and activation of the mTOR pathway, as measured by phosphorylation of protein S6 (48). Loss of arid1a in mouse pancreas leads to acinar to ductal metaplasia and, when associated with KRAS and TP53 mutations, to development of cancer with short latency and poor differentiation (49). This loss promotes epithelial-mesenchymal transition in human pancreatic cells and sensitizes human pancreatic cancer xenografts to treatment with HSP90 inhibitor NVP-AUY922, by destabilizing client protein vimentin, which is upregulated in ARID1A null cells (50). Decreased activity of ARID1A is required for development of pre-cancerous lesions in the pancreas, including pancreatic intraepithelial neoplasia, intraductal tubulopapillary neoplasm and IPMN (51). ARID1A as part of the SWI/SNF complex participates in DNA repair, interacting with kinase ATR and promoting ATR activation in double strand breaks and induction of the G2/M DNA damage checkpoint (46). Cancer cells with ARID1A deficiency are sensitive to PARP inhibitors olaparib, veliparib, rucaparib and BMN673 (52). Moreover, a case report of a metastatic pancreatic cancer patient with a pathogenic ARID1A mutation (and concomitant KRAS and TP53 mutations in this case) showed a response lasting for over a year (53). Pancreatic cancer patients with SWI/SNF mutations, such as ARID1A, ARID1B, SMARCA4 and SMARCB1, are reported to benefit from immunotherapy with checkpoint inhibitors, independently from their MSI status, a finding awaiting prospective validation (54).
BCOR (BCL6 transcription corepressor), another epigenetic modifier and a member of Polycomb Repressive Complex 1 (PRC1), is mutated in a small percentage of pancreatic cancers without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations. Mutations in BCOR and related PRC1 subunit BCORL1 are involved in leukemogenesis, leading to dysfunction of PRC1 and oncogenic signaling through derepression of normally suppressed genes (55). Although this molecular defect is not currently targeted directly, data from lung cancer suggest that BCOR mutations as well as mutations in two other epigenetic modifiers, KMT2C and KDM5C, may be predictive of response to immune checkpoint inhibitors (56).
Fusions involving NRG1, the gene encoding for the HER3 ligand Neuregulin 1, are observed in a subset of cases without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations. These and a few other fusions are enriched in cases without the common pancreatic cancer molecular alterations compared with pancreatic cancers presenting with these alterations. NRG1 fusions were previously reported in pancreatic cancer patients without KRAS mutations and are present in other cancers, including lung, ovarian and endometrial cancers (57,58). The partner in these fusions varies but the NRG1 part consistently contains the EGF-like domain of the protein, which enables HER3 binding and oncogenic signaling downstream through the KRAS/BRAF/MEK and the PI3K/AKT pathways (59). Thus, NRG1 fusion containing pancreatic cancers possess an alternative way of activating KRAS (60). Targeting with anti-HER3 monoclonal antibodies in development may offer a viable therapeutic option in this subset of molecularly defined pancreatic cancers (61-63). Other rare fusions that are present in pancreatic cancers with or without common alterations present targeted therapeutic opportunities (63).
Studies based on genomic profiles of pancreatic adenocarcinomas have shown that, similar to other cancers, the disease is heterogeneous and can be classified into four sub-types: squamous, pancreatic progenitor, aberrant differentiation endocrine-exocrine (ADEX) and immunogenic (64). The squamous sub-type is characterized by mutations in TP53 and KDM6A and down-regulation of endodermal cell fate determiner genes PDX1, MNX1, GATA6 and HNF1B (64). This sub-type corresponds to the quasi-mesenchymal sub-type of another genomic classification proposed by Collisson et al. (65). In contrast, the pancreatic progenitor sub-type, corresponding to the classical sub-type of Collisson et al., shows up-regulation of PDX1, MNX1 and HNF1B as well as of HNF4G, HNF4A, HNF1A, FOXA2, FOXA3 and HES1 (64). The ADEX subtype, corresponding to the exocrine-like sub-type of Collisson et al., is characterized by both up-regulation of exocrine transcription factors such as NR5A2, BHLH1A15 and RBPJL and up-regulation of endocrine differentiation markers such as INS, NEUROD1, NKX2-2 and MAFA. Several cases in the group of pancreatic cancers without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations in TCGA cohort show up-regulation of these exocrine and endocrine defining genes, suggesting that they align with the ADEX sub-type. In addition, a few cases have immune checkpoint proteins up-regulation suggesting that they belong to the immunogenic sub-type. Genomic classifications have yet to be validated as predictive markers of therapies and their clinical value remains to be determined (66). However, they may imply different pathogenesis of sub-types, which could facilitate successful rational targeted therapies development. Thus, membership of cases without common alterations in the ADEX or immunogenic sub-type may allow their inclusion in therapeutic trials aiming at these groups.
OS of the group of cancers without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations in TCGA cohort is better than OS of counterparts with common alterations (Log Rank P=0.03, Figure 3). In contrast, the OS comparison between the two groups in the MSK-IMPACT series, disclosed no differences (Log Rank P=0.4, not shown). This inconsistency may be due to differences in the stage of included cases as the group without KRAS, TP53, SMAD4, CDKN2A and CDKN2B alterations in TCGA contained a significantly higher percentage of stage I patients, while the two groups in the MSK-IMPACT cohort which showed no survival difference, had no significant different stages distribution. Alternatively, absence of common alterations may have different prognostic implications according to the stage of the disease. Indeed, absence of the most common KRAS G12D mutations were not prognostic in a cohort of pancreatic cancer patients across stages, but were associated with a better survival compared with patients that had mutated cancers in localized resectable disease (67).
Conclusions
In conclusion, the sub-set of pancreatic cancers without the common molecular alterations characterizing the disease has a prevalence that is somewhat variable between series but is between 5% and 10% in most series. Although their prognosis stage by stage may not be distinct from cases with common alterations, these cases are a group still possessing molecular lesions of therapeutic interest. These alterations could serve as therapeutic targets, as additional molecularly based therapies are introduced in the clinic.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://cco.amegroups.com/article/view/10.21037/cco-22-108/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Arnold M, Abnet CC, Neale RE, et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology 2020;159:335-349.e15. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Conroy T, Desseigne F, Ychou M, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817-25. [Crossref] [PubMed]
- Conroy T, Hammel P, Hebbar M, et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N Engl J Med 2018;379:2395-406. [Crossref] [PubMed]
- Thibodeau S, Voutsadakis IA. FOLFIRINOX Chemotherapy in Metastatic Pancreatic Cancer: A Systematic Review and Meta-Analysis of Retrospective and Phase II Studies. J Clin Med 2018;7:7. [Crossref] [PubMed]
- Leal AD, Messersmith WA, Lieu CH. Neoadjuvant treatment of localized pancreatic adenocarcinoma. J Gastrointest Oncol 2021;12:2461-74. [Crossref] [PubMed]
- Lambert A, Conroy T, Ducreux M. Future directions in drug development in pancreatic cancer. Semin Oncol 2021;48:47-56. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard; . Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32:185-203.e13. [Crossref]
- Lowery MA, Jordan EJ, Basturk O, et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res 2017;23:6094-100. [Crossref] [PubMed]
- Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703-13. [Crossref] [PubMed]
- Xu S, Guo Y, Zeng Y, et al. Clinically significant genomic alterations in the Chinese and Western patients with intrahepatic cholangiocarcinoma. BMC Cancer 2021;21:152. [Crossref] [PubMed]
- AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 2017;7:818-31. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [Crossref] [PubMed]
- Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [Crossref] [PubMed]
- Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018;33:676-689.e3. [Crossref] [PubMed]
- Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413-21. [Crossref] [PubMed]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [Crossref] [PubMed]
- Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 2015;17:251-64. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017:PO.17.00011.
- Luo J. KRAS mutation in pancreatic cancer. Semin Oncol 2021;48:10-8. [Crossref] [PubMed]
- Thompson ED, Roberts NJ, Wood LD, et al. The genetics of ductal adenocarcinoma of the pancreas in the year 2020: dramatic progress, but far to go. Mod Pathol 2020;33:2544-63. [Crossref] [PubMed]
- Bailey JM, Hendley AM, Lafaro KJ, et al. p53 mutations cooperate with oncogenic Kras to promote adenocarcinoma from pancreatic ductal cells. Oncogene 2016;35:4282-8. [Crossref] [PubMed]
- Voutsadakis IA. Mutations of p53 associated with pancreatic cancer and therapeutic implications. Ann Hepatobiliary Pancreat Surg 2021;25:315-27. [Crossref] [PubMed]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321:1801-6. [Crossref] [PubMed]
- Strickler JH, Satake H, George TJ, et al. Sotorasib in KRAS p.G12C-Mutated Advanced Pancreatic Cancer. N Engl J Med 2023;388:33-43. [Crossref] [PubMed]
- Bannoura SF, Khan HY, Azmi AS. KRAS G12D targeted therapies for pancreatic cancer: Has the fortress been conquered? Front Oncol 2022;12:1013902. [Crossref] [PubMed]
- Philip PA, Azar I, Xiu J, et al. Molecular Characterization of KRAS Wild-type Tumors in Patients with Pancreatic Adenocarcinoma. Clin Cancer Res 2022;28:2704-14. [Crossref] [PubMed]
- Hsu FC, Roberts NJ, Childs E, et al. Risk of Pancreatic Cancer Among Individuals With Pathogenic Variants in the ATM Gene. JAMA Oncol 2021;7:1664-8. [Crossref] [PubMed]
- Jette NR, Kumar M, Radhamani S, et al. ATM-Deficient Cancers Provide New Opportunities for Precision Oncology. Cancers (Basel) 2020;12:687. [Crossref] [PubMed]
- Lloyd RL, Wijnhoven PWG, Ramos-Montoya A, et al. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene 2020;39:4869-83. [Crossref] [PubMed]
- Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol Cell 2017;66:801-17. [Crossref] [PubMed]
- Gout J, Perkhofer L, Morawe M, et al. Synergistic targeting and resistance to PARP inhibition in DNA damage repair-deficient pancreatic cancer. Gut 2021;70:743-60. [Crossref] [PubMed]
- Roger E, Gout J, Arnold F, et al. Maintenance Therapy for ATM-Deficient Pancreatic Cancer by Multiple DNA Damage Response Interferences after Platinum-Based Chemotherapy. Cells 2020;9:2110. [Crossref] [PubMed]
- Golan T, Kanji ZS, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111:1132-8. [Crossref] [PubMed]
- Momtaz P, O'Connor CA, Chou JF, et al. Pancreas cancer and BRCA: A critical subset of patients with improving therapeutic outcomes. Cancer 2021;127:4393-402. [Crossref] [PubMed]
- Golan T, Barenboim A, Lahat G, et al. Increased Rate of Complete Pathologic Response After Neoadjuvant FOLFIRINOX for BRCA Mutation Carriers with Borderline Resectable Pancreatic Cancer. Ann Surg Oncol 2020;27:3963-70. [Crossref] [PubMed]
- More A, Ito I, Haridas V, et al. Oncogene addiction to GNAS in GNAS(R201) mutant tumors. Oncogene 2022;41:4159-68. [Crossref] [PubMed]
- Mas L, Lupinacci RM, Cros J, et al. Intraductal Papillary Mucinous Carcinoma Versus Conventional Pancreatic Ductal Adenocarcinoma: A Comprehensive Review of Clinical-Pathological Features, Outcomes, and Molecular Insights. Int J Mol Sci 2021;22:6756. [Crossref] [PubMed]
- Hosoda W, Sasaki E, Murakami Y, et al. GNAS mutation is a frequent event in pancreatic intraductal papillary mucinous neoplasms and associated adenocarcinomas. Virchows Arch 2015;466:665-74. [Crossref] [PubMed]
- Omori Y, Ono Y, Tanino M, et al. Pathways of Progression From Intraductal Papillary Mucinous Neoplasm to Pancreatic Ductal Adenocarcinoma Based on Molecular Features. Gastroenterology 2019;156:647-661.e2. [Crossref] [PubMed]
- Sohn TA, Yeo CJ, Cameron JL, et al. Intraductal papillary mucinous neoplasms of the pancreas: an updated experience. Ann Surg 2004;239:788-97; discussion 797-9. [Crossref] [PubMed]
- Tan MC, Basturk O, Brannon AR, et al. GNAS and KRAS Mutations Define Separate Progression Pathways in Intraductal Papillary Mucinous Neoplasm-Associated Carcinoma. J Am Coll Surg 2015;220:845-854.e1. [Crossref] [PubMed]
- Kawabata H, Ono Y, Tamamura N, et al. Mutant GNAS limits tumor aggressiveness in established pancreatic cancer via antagonizing the KRAS-pathway. J Gastroenterol 2022;57:208-20. [Crossref] [PubMed]
- Luchini C, Brosens LAA, Wood LD, et al. Comprehensive characterisation of pancreatic ductal adenocarcinoma with microsatellite instability: histology, molecular pathology and clinical implications. Gut 2021;70:148-56. [Crossref] [PubMed]
- Kimura Y, Fukuda A, Ogawa S, et al. ARID1A Maintains Differentiation of Pancreatic Ductal Cells and Inhibits Development of Pancreatic Ductal Adenocarcinoma in Mice. Gastroenterology 2018;155:194-209.e2. [Crossref] [PubMed]
- Wang W, Friedland SC, Guo B, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut 2019;68:1245-58. [Crossref] [PubMed]
- Tomihara H, Carbone F, Perelli L, et al. Loss of ARID1A Promotes Epithelial-Mesenchymal Transition and Sensitizes Pancreatic Tumors to Proteotoxic Stress. Cancer Res 2021;81:332-43. [Crossref] [PubMed]
- Liu M. Arid1a: A Gatekeeper in the Development of Pancreatic Cancer From a Rare Precursor Lesion. Gastroenterology 2022;163:371-3. [Crossref] [PubMed]
- Shen J, Peng Y, Wei L, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint and Sensitizes Cells to PARP Inhibitors. Cancer Discov 2015;5:752-67. [Crossref] [PubMed]
- Zhao XS, Zhou J, Dong L, et al. Durable response to olaparib in pancreatic duct adenocarcinoma with deleterious ARID1A mutation. Chin Med J (Engl) 2019;132:3012-4. [Crossref] [PubMed]
- Botta GP, Kato S, Patel H, et al. SWI/SNF complex alterations as a biomarker of immunotherapy efficacy in pancreatic cancer. JCI Insight 2021;6:e150453. [Crossref] [PubMed]
- Schaefer EJ, Wang HC, Karp HQ, et al. BCOR and BCORL1 Mutations Drive Epigenetic Reprogramming and Oncogenic Signaling by Unlinking PRC1.1 from Target Genes. Blood Cancer Discov 2022;3:116-35. [Crossref] [PubMed]
- Liu D, Benzaquen J, Morris LGT, et al. Mutations in KMT2C, BCOR and KDM5C Predict Response to Immune Checkpoint Blockade Therapy in Non-Small Cell Lung Cancer. Cancers (Basel) 2022;14:2816. [Crossref] [PubMed]
- Jones MR, Williamson LM, Topham JT, et al. NRG1 Gene Fusions Are Recurrent, Clinically Actionable Gene Rearrangements in KRAS Wild-Type Pancreatic Ductal Adenocarcinoma. Clin Cancer Res 2019;25:4674-81. [Crossref] [PubMed]
- Jones MR, Lim H, Shen Y, et al. Successful targeting of the NRG1 pathway indicates novel treatment strategy for metastatic cancer. Ann Oncol 2017;28:3092-7. [Crossref] [PubMed]
- Aguirre AJ. Oncogenic NRG1 Fusions: A New Hope for Targeted Therapy in Pancreatic Cancer. Clin Cancer Res 2019;25:4589-91. [Crossref] [PubMed]
- Zhang C, Mei W, Zeng C. Oncogenic Neuregulin 1 gene (NRG1) fusions in cancer: A potential new therapeutic opportunities. Biochim Biophys Acta Rev Cancer 2022;1877:188707. [Crossref] [PubMed]
- Drilon A, Somwar R, Mangatt BP, et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer Discov 2018;8:686-95. [Crossref] [PubMed]
- Umemoto K, Sunakawa Y. The potential targeted drugs for fusion genes including NRG1 in pancreatic cancer. Crit Rev Oncol Hematol 2021;166:103465. [Crossref] [PubMed]
- Zheng-Lin B, O'Reilly EM. Pancreatic ductal adenocarcinoma in the era of precision medicine. Semin Oncol 2021;48:19-33. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Schreyer D, Neoptolemos JP, Barry ST, et al. Deconstructing Pancreatic Cancer Using Next Generation-Omic Technologies-From Discovery to Knowledge-Guided Platforms for Better Patient Management. Front Cell Dev Biol 2022;9:795735. [Crossref] [PubMed]
- Shen H, Lundy J, Strickland AH, et al. KRAS G12D Mutation Subtype in Pancreatic Ductal Adenocarcinoma: Does It Influence Prognosis or Stage of Disease at Presentation? Cells 2022;11:3175. [Crossref] [PubMed]


