Genetic and epigenetic landscape of nasopharyngeal carcinoma
Introduction
Nasopharyngeal carcinoma (NPC) is a unique malignancy that arises from the epithelium of the nasopharynx and has a restricted prevalence in certain regions of the world. NPC shows the highest incidence rate in the Southeast China, Southeast Asia, North Africa, Middle East, and the Arctic regions, but it is rare in most other parts of the world (1). NPC patients are relatively young at diagnosis, when compared to other cancer types that usually are diagnosed at 65 years of age or older, while NPC has a median age at diagnosis of around 50.
Previous studies suggested that there are several cofactors that are associated with NPC development. They are the dietary factors, infection with an oncovirus, Epstein-Barr virus (EBV), and genetic predisposition. NPC is associated with genetic alterations on particular chromosomal regions and genes, harboring of specific cancer-associating single nucleotide polymorphisms (SNPs), and familial aggregation. Interestingly, recent studies confirm that epigenetic alterations, including the promoter hypermethylation (2), are also one of the crucial factors that are highly associated with NPC.
Previous reviews suggested the multi-step progression model for NPC. Lo et al. proposed that the loss of chromosomes 3p and 9p regions is an early event for the transformation of the normal nasopharyngeal epithelium (3). During the transformation process, the inactivation of the p16 gene on chromosome 9 and RASSF1 on chromosome 3, and/or the over-expression of the cyclin D1 or amplification of particular regions on chromosome 11 are followed by the EBV latent infection and multiple genetic and epigenetic changes. The subsequent loss of chromosomes 11q, 13q, 14q, and 16q regions and promoter hypermethylation of numerous tumor suppressor genes (TSGs), including E-cadherin, finally result in NPC (3). This model proposes a critical role of the genetic and epigenetic factors in the pathogenesis of NPC. In this review, we summarize the genetic and epigenetic changes in NPC, as seen in Figure 1, and focus discussions on these critical events that contribute to the initiation and progression of this EBV-associated tumor, as well as the potential use of these findings as biomarkers for NPC diagnosis and prognosis.
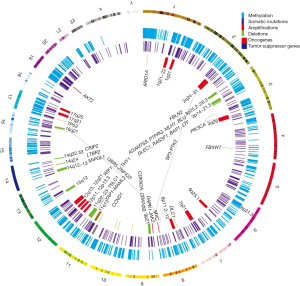
Genetic alterations in NPC
In 2014, the first genomic landscape study of NPC was published using the whole-exome sequencing (WES), targeted deep sequencing, and SNP array analysis approaches to characterize NPC genetic alterations (4). This integrated study revealed the enrichment of genetic lesions affecting several important cellular processes and pathways including chromatin modification, ERBB-PI3K signaling, and autophagy machinery.
Somatic mutations
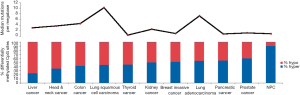
The somatic mutation rate in NPC is relatively low compared to other types of cancers, with a median of one somatic mutation per megabase (Figure 2). Interestingly, TP53 is the most frequently mutated gene in NPC. Most of the earlier studies using the single-strand conformation polymorphism (SSCP) method to screen for mutations reported infrequent TP53 mutations in NPC biopsies (6,7). Using a sensitive yeast screening assay developed by Flaman et al. (8), that covers both the exons and introns of p53, Lung et al. (9) were able to detect up to 27% mutated p53 in a small number of NPC biopsies, indicating that the SSCP methods were inadequate to detect many of the p53 mutations harbored in NPC. Now with sensitive WES approaches, higher TP53 mutation frequencies of 8.5% were observed in a Singapore study (4), as well as in 7.3% of our NPC biopsies in a Hong Kong study (manuscript in revision).
Most well-known oncogenes are rarely mutated in NPC. Mutation hotspots in EGFR, PIK3CA, KRAS, HRAS, NRAS, BRAF, KIT, ABL1, AKT1, AKT2, CDK4, ERBB2, FGFR1, FGFR3, FLT3, JAK2, MET, RET, and PDGFRA were screened using SNaPshot or OncoCarta panel assays in 70 and 160 NPC patients, respectively. In each study, most genes were mutated in less than three NPC samples at the mutation hotspots in each study, except for KIT, which was reported to be mutated in 10% (7/70) and 2% (3/160) of NPC samples (10,11). WES identified mutations in multiple members of the ERBB-PI3K pathway including PIK3CA (1.7%), ERBB2 (1.7%), ERBB3 (1.7%), FGFR2 (2.6%), KRAS (1.7%), and NRAS (0.9%) (4). In another study PIK3CA was shown to be mutated in 9.6% (7/73) of NPC cases using polymerase chain reaction (PCR)-sequencing methods (12).
Both the published WES study (4), as well as our WES data, show that epigenetic regulators are frequently mutated in NPC, including ARID1A, BAP1, KMT2B, KMT2C, KMT2D, TSHZ3, HDAC4, PAXIP1, and others. Other mutated genes identified by WES include cadherin members (FAT1, FAT2, FAT3), Notch family members (NOTCH1, NOTCH2, NOTCH3), and autophagy-related genes (ATG2A, ATG7, ATG13) (4).
Copy number gains
Multiple chromosomal abnormalities and gene copy number alterations have been identified in NPC by various approaches including comparative genomic hybridization (CGH), microsatellite marker analysis, karyotyping, array-based CGH (aCGH), and SNP array. Chromosomes 1q, 2q, 3q, 4q, 6q, 7q, 8p, 8q, 11q, 12p, 12q, and 17q frequently have copy number gains in NPC (13-17). Cyclin D1 (CCND1), located at 11q13, has the highest frequency of copy number changes in NPC, with a reported frequency ranging from 9.6% (5/52 using SNP array) to 61.9% (13/21 using aCGH) (4,18). Another oncogene related to G1/S transition, MYC, located in 8q24, was also amplified in 5.8% (3/52 by SNP array) to 62% (21/34 by FISH) of NPC tumors (4,19).
Multiple members of the ERBB-PI3K pathway also have copy number gains apart from somatic mutations. SNP array and aCGH analysis revealed amplifications in PIK3CA (75%, 24/32 with aCGH and FISH), AKT2 (5.8%, 3/52 with SNP array), and JAK2 (3.8%, 2/52 with SNP array) (4,20). Mutations and amplifications were frequently observed in the ERBB-PI3K pathway regulators, indicating the importance of this pathway in NPC.
Other frequent amplifications identified by CGH include LTBR at 12p13, TERC at 3q26.3, and ESR at 6q25.1 (21,22).
Copy number losses and loss of heterozygosity (LOH)
Previous studies suggested the loss of chromosomes 1p, 3p, 9p, 9q, 11q, 13q, 14q, and 16q regions are common events in NPC (13-17,23-25). Functional complementation studies were performed by utilizing the microcell-mediated chromosome transfer (MMCT) approach to transfer intact or truncated chromosomes to identify the critical regions (CRs) that are important for tumor suppression in NPC cells, including 3p21.3, 14q12, 14q13.2–13.3, 14q24.1, 14q32.33, 11q13, and 11q22–23 (26-32). The 3p21.3 region contains several important TSGs. BLU/ZMYND10, PTPRG, RASSF1A, and FBLN2, mapped to or next to this CR, were identified by MMCT, cDNA microarray assay, tissue microarray staining, and NotI chromosome 3 genomic microarray analysis (33-36). Other candidate TSGs located in 3p include ADAMTS9, CACNA2D3, GNAT1, LTF, LARS2, RBMS3, and CHL1 (13,37). Several TSGs on chromosome 14, including the MIPOL1, CRIP2, and LTBP2, showed significant in vitro and in vivo tumor-suppressive effects (26,38-40). CRs on chromosome 11, 11q13, and 11q22–23, harbored TSGs including TSLC1/CADM1 (41), THY1 (42), and CRYAB (27).
Frequent chromosome 9p losses were found in histologically normal nasopharyngeal epithelia and, thus, were considered as a critical early event in NPC tumorigenesis (23). Homozygous deletions of the 9p21.3 region containing CDKN2A (p16) and CDKN2B (p15) were found in NPC xenografts and primary tumors (43). Consistently, the genomic landscape study showed the most frequent deletion occurs in the 9p21 region spanning CDKN2A and CDKN2B (4).
Apart from somatic mutations, SNP array analysis revealed several chromatin modification regulators that also harbor frequent number losses in NPC, including ARID1A (15.4%), BAP1 (1.9%), and TET1 (3.8%), which further emphasized the importance of this pathway. TP53, which is the most frequently mutated gene in NPC, was also reported to be deleted in 15.4% of NPC cases (8/52) (4).
Gene translocations and rearrangements
Only a few structural variants were reported in NPC. A fusion of ubiquitin protein ligase E3 component n-recognin 5 (UBR5) on 8q22.3 and zinc finger protein 423 (ZNF423) on 16q12.1 was first identified by whole-transcriptome sequencing from the C666 cell line and later was recurrently detected in 8.3% (12/144) of primary tumors (44). Functional analysis demonstrated that the fusion protein affected cell proliferation and colony-forming ability in vitro and promoted tumor formation in vivo (44). Another study developed a new algorithm to detect structural variants from high-throughput sequencing from long-insert paired-end DNA libraries of NPC, and identified gene fusions at YAP1-MAML2, PTPLB-RSRC1, and SP3-PTK2. FISH analysis of an additional 196 cases identified more rearrangements at MAML2 (two cases), PTK2 (six cases), and SP3 (three cases) (45).
Epigenetic alterations in NPC
Multiple factors including host genetics, viral infection, and environmental factors contribute to NPC tumorigenesis, which all play integral roles in modulating the epigenetic changes, ultimately leading to the critical gene expression alterations. Aberrant epigenetic alterations, particularly DNA methylation, have been frequently reported in NPC. In our recent comparative methylome study, we found NPC had the highest hypermethylation frequency, as compared to nine other cancer types including liver, head and neck, colon, lung, thyroid, kidney, breast, pancreatic, and prostate cancers (2) (Figure 2). A number of tumor suppressors were found to be downregulated by promoter methylation. Aberrant methylation of tumor suppressors RASSF1 and CDKN2A was considered as important early events in NPC tumorigenesis. Promoter methylation of RASSF1 and CDKN2A was found in 49–83% and 16–66% of NPC patients, respectively. Moreover, we identified a number of novel tumor suppressors including ADAMTS9 (37), PTPRG (34), ZMYND10 (33), FBLN2 (35), CRYAB (27), CADM1 (41), THY1 (42), MMP19 (46), DUSP6 (47), MIPOL1 (26), and LTBP2 (39) by MMCT studies in NPC; the majority of these tumor suppressors were downregulated in NPC by promoter methylation (48). Important DNA methylation changes are frequently reported in several CRs such as 3p21.3, 9p21, and 6p21.3 regions as summarized in Table 1.
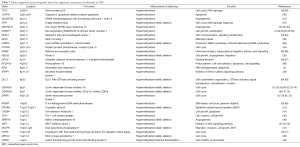
Full table
3p21.3 region
Promoter methylation at multiple 3p21.3 genes has been reported in NPC. The most important epigenetic event was RASSF1 promoter methylation, which was originally reported in NPC xenografts, cell lines, and in primary tumors, but not in the normal nasopharyngeal epithelia (55). These results were further confirmed by several studies carried out at the single gene or genome-wide levels (2,49,50,52,55-63), while somatic mutations at this gene were detected at only low frequencies (9.5%) in primary tumors and were not observed in recent somatic landscape studies with the Singapore cohort (4) and in our Hong Kong study (manuscript in revision). Besides RASSF1, promoter methylation was also detected in MLH1 (50), DLEC1 (53,64), LARS2 (65), SLC6A20 (2), LTF (2), and ZMYND10 (BLU) (52-54) from the 3p21.3 region in NPC patients, suggesting DNA methylation plays an important role in regulating the expression of the genes in this region.
6p22.1−6p21.3 region
The human major histocompatibility complex (MHC) locus is located at the 6p22.1–6p21.3 region. In NPC, the association between human leukocyte antigen (HLA) polymorphisms with NPC genetic susceptibility was confirmed by genome-wide association study (GWAS). Interestingly, the methylome study using the Illumina Infinium assay on HumanMethylation450 BeadChip reveals significant methylation changes in this region in NPC (2). A number of genes including B3GALT4, FLOT1, PXT1, TNXB, TRIM31, LY6G5C, PPP1R18, GNL1, IER3, and NKAPL in this region were hypermethylated in tumors compared to non-tumor tissues, indicating the importance of DNA methylation in regulating gene expression. Although none of the MHC class I HLA genes was found significantly methylated in NPC, previous study using gene expression profiling microarrays showed the inhibition of the expression of the MHC class I genes in NPC by EBV (78). It is likely that epigenetic changes have a role in regulating the transcription of HLA genes in NPC. Further investigation of aberrant methylation in this region in a larger patient cohort is necessary. Although next-generation sequencing (NGS)-based bisulfite sequencing has emerged as a powerful method to characterize the methylation changes in high resolution, MHC is a highly polymorphic region, which limits the utility of NGS-based methods, since the complexity of the DNA sequence is greatly reduced after bisulfite conversion. An alignment and methylation analysis pipeline specifically designed for the MHC region having adequate sequencing depth is required to improve the accuracy of methylation assessment in this region.
9p21 region
Homozygous deletion of the 9p21.3 region containing CDKN2A (p16) and CDKN2B (p15) was found in NPC. In addition to genomic alterations, inactivation of both genes by promoter methylation was reported in 16–66% and 21–50% of the tumors, respectively (50,53,58,60,62). These results emphasize the importance of integrating the genetic and epigenetic changes to provide a comprehensive view of molecular changes in NPC.
Signaling pathways deregulated by DNA methylation in NPC
Two genome-wide methylome studies consistently identified a few important signaling pathways and functions often deregulated by DNA methylation in NPC, including the Wnt, MAPK, Hedgehog, and TGF-β signaling pathways and focal adhesion (2,49). In the Wnt signaling pathway, a number of Wnt inhibitors including DKK1, WIF1, SFRP1, SFRP2, SFRP4, and SFRP5 are silenced by promoter methylation in NPC. Inactivation of these inhibitors may lead to the aberrant activation of Wnt signaling and transcription of its downstream targets. Gene expression profiling of primary tumors shows the differential expression of several components of the Wnt canonical signaling pathway (79). These data suggested the important involvement of aberrant Wnt signaling in NPC tumorigenesis.
Methylation status as biomarkers for NPC diagnosis and prognosis
Given the widespread aberrant methylation often observed in NPC, we and others have evaluated the usefulness of methylation biomarker panels for NPC early detection, using tumor biopsies and non-invasive biological specimens as summarized in Table 2 (57-60,62,64,70-72). For example, Wong and his colleagues examined the methylation of CDH1, DAPK1, CDKN2B, RASSF1, and CDKN2A in plasma samples from 41 NPC patients before treatment and 43 normal individuals by real-time quantitative PCR after bisulfite conversion; they found aberrant methylation in at least one of the five genes was detectable in 71% plasma of NPC patients with a specificity of 91% (72). Methylation of five tumor suppressors (RASSF1, CDKN2A, DLEC1, DAPK1, UCHL1) in serum samples was investigated in 40 NPC patients and 41 age- and gender-matched healthy subjects using the methylation-specific PCR approach (58). Combination of four markers including CDKN2A, DLEC1, DAPK1, and UCHL1 has the highest sensitivity and specificity for detecting NPC. Circulating EBV copy number changes have emerged as a promising biomarker to aid NPC diagnosis (80). We examined a methylation panel including RASSF1, WIF1, DAPK1, and RARB2 using high-resolution melting curve analysis and EBV copy number changes in 220 plasma samples from NPC patients and found this methylation panel can be used as a supplementary test to the EBV copy number test for increased detection of early-stage NPC (57). Combinations of both the four-gene methylation panel and EBV DNA test can detect 63.6%, 86.7%, 88%, and 96.5% of the patients with stages I, II, III, and IV tumors, respectively, with a specificity of 88%. In addition to hypermethylation mainly at promoter CpG islands, global hypomethylation predominantly in the repetitive elements and gene body, is another feature of cancer. Chan et al. explored the usage of genome-wide hypomethylation and copy number aberrations in plasma DNA by bisulfite sequencing in multiple types of cancers including NPC with a sensitivity of 74% and a specificity of 94% (81). However, this NGS-based method is costly and further evaluation in a larger patient cohort is necessary prior to clinical use.

Full table
NPC local recurrence and distant metastasis contribute to treatment failure and NPC mortality. Therefore, identification of biomarkers for detecting the recurrence and metastasis and predicting the patient prognosis is extremely important for improving the treatment success in NPC. In our methylation biomarker study carried out in plasma samples, a combination of the four-gene methylation panel and EBV DNA tests has a sensitivity as high as 94.3% and 93.5% for detection of the metastasis and local recurrence, respectively, suggesting the combination testing has great potential for monitoring disease progression. Recently, a genome-wide methylation study identified a panel of prognostic methylation biomarkers comprising six hypermethylated genes WIF1, CCNA1, UCHL1, RASSF1, TP73, and SFRP1 (56). The study was carried out in two stages including the training and validation cohorts. In the training cohort (n=150), the patients with high methylation levels had poorer disease-free survival (DFS) [HR, 2.26; 95% confidence interval (CI), 1.28–4.01; P=0.005] and overall survival (OS) [HR, 2.47; 95% CI, 1.30–4.71; P=0.006] than those with low methylation. The association of the methylation panel with survival was independent of stage. The prognostic value was further validated in two independent cohorts including 150 and 153 NPC patients, respectively. The patients with low methylation had a favorable response to concurrent chemotherapy with an improved DFS and OS, whereas patients with high methylation did not benefit a present chemotherapy. This study demonstrated the potential usefulness of a methylation panel as a prognostic and predictive tool in NPC.
Aberrant histone modifications in NPC
In the genomic landscape study in NPC, somatic mutations were identified in a number of histone modifiers, suggesting the importance of aberrant histone modification in NPC tumorigenesis. Consistently, our methylome study reveals that the aberrantly methylated genes often have bivalent markers, H3K4me3 mark for active transcription and H3K27me3 for repressive transcription, indicating DNA methylation functions together with histone modification for regulating gene expression in NPC tumorigenesis (2). The enrichment of the genes with homeobox domain in the aberrantly methylated genes in NPC indicates these aberrantly methylated genes are often polycomb complex targets. Many studies have demonstrated that the polycomb repressive complex genes encoding epigenetic gene-silencing proteins contribute to the oncogenesis of various cancers. Over-expression of the polycomb complex genes BIM1 and EZH2 was demonstrated in NPC tumors (82,83). Importantly, ectopic expression of BMI1 is sufficient to immortalize the normal nasopharyngeal epithelial (NPE) cells (83). EZH2 encodes a histone-lysine N-methyltransferase, responsible for H3K27 methylation. The important roles of EZH2 in NPC cell migration/invasion, cell survival, and angiogenesis have been demonstrated in vitro in several studies (84-86). Cai et al. examined the H3K27me3 level in the NPC tumors and normal NPE tissues by Western blotting and immunohistochemistry (IHC). They found 60.8% of NPC and only 16% of normal NPE tissues have high level of H3K27me3 marks and the H3K27me3 level is associated with patient prognosis and chemoradiotherapy response (87).
EBV and epigenetic alterations
In NPC, EBV is harbored in a latent stage with restricted viral gene expression of LMP1/2, EBNA1, EBER1/2, and BARTs. LMP1 can activate cellular DNA methyltransferase via c-Jun NH2-terminal kinase signaling (88) and upregulate BMI1 expression (89), which is associated with epigenetic changes in NPC. On the other hand, latent episomal EBV genomes are subject to host cell-dependent epigenetic modifications including DNA methylation and histone modifications, which are important for maintenance of EBV latent infection (90).
In addition to NPC, extensive methylation changes are also often observed in the EBV-associated gastric cancer (91). Previous studies suggested that the EBV-infected cells acquire extensive methylation to silence multiple TSGs and to transform cells into cancer cells (92). Although the initial step of virus infection in the epithelial cell and the methylation mechanism induced by EBV infection has not been fully elucidated, it was proposed that EBV infection is an epigenetic driver in tumorigenesis (92).
Conclusions and future directions
NPC is a complex disease that involves host genetics, viral infection, and environmental factors. It is characterized by a comparatively low mutation rate, extensive hypermethylation, as well as frequent chromosomal abnormalities and copy number alterations. Although the high-throughput characterization methods, such as massive parallel sequencing, SNP, and methylation assays, has resulted in a greater understanding of NPC biology, yet much remains to be uncovered. Moreover, integration of the whole-exome and whole-genome sequencing data, transcriptome, proteomics, and methylome data in more NPC patients, together with well-documented clinical outcome information, will aid us to further understand the importance of molecular alterations and their clinical relevance. Study of additional NPC patients will enhance our ability to identify the less frequent, but critical, genetic and epigenetic events in the pathogenesis of NPC. These insights will lead to the identification of powerful biomarkers for NPC diagnosis and prognosis and evaluation of novel and existing targeted therapies to improve the treatment success.
Acknowledgements
Funding: This study was funded by the Hong Kong Research Grants Council grant AoE/M-06/08 to MLL.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Yu MC, Yuan JM. Epidemiology of nasopharyngeal carcinoma. Semin Cancer Biol 2002;12:421-9. [Crossref] [PubMed]
- Dai W, Cheung AK, Ko JM, et al. Comparative methylome analysis in solid tumors reveals aberrant methylation at chromosome 6p in nasopharyngeal carcinoma. Cancer Med 2015;4:1079-90. [Crossref] [PubMed]
- Lo KW, Chung GT, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Semin Cancer Biol 2012;22:79-86. [Crossref] [PubMed]
- Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet 2014;46:866-71. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Lo KW, Mok CH, Huang DP, et al. p53 mutation in human nasopharyngeal carcinomas. Anticancer Res 1992;12:1957-63. [PubMed]
- Spruck CH 3rd, Tsai YC, Huang DP, et al. Absence of p53 gene mutations in primary nasopharyngeal carcinomas. Cancer Res 1992;52:4787-90. [PubMed]
- Flaman JM, Frebourg T, Moreau V, et al. A simple p53 functional assay for screening cell lines, blood, and tumors. Proc Natl Acad Sci U S A 1995;92:3963-7. [Crossref] [PubMed]
- Lung ML, Hu Y, Cheng Y, et al. p53 inactivating mutations in Chinese nasopharyngeal carcinomas. Cancer Lett 1998;133:89-94. [Crossref] [PubMed]
- Zhang JW, Qin T, Hong SD, et al. Multiple oncogenic mutations related to targeted therapy in nasopharyngeal carcinoma. Chin J Cancer 2015;34:177-83. [Crossref] [PubMed]
- Jiang N, Liu N, Yang F, et al. Hotspot mutations in common oncogenes are infrequent in nasopharyngeal carcinoma. Oncol Rep 2014;32:1661-9. [PubMed]
- Chou CC, Chou MJ, Tzen CY. PIK3CA mutation occurs in nasopharyngeal carcinoma but does not significantly influence the disease-specific survival. Med Oncol 2009;26:322-6. [Crossref] [PubMed]
- Chen J, Fu L, Zhang LY, et al. Tumor suppressor genes on frequently deleted chromosome 3p in nasopharyngeal carcinoma. Chin J Cancer 2012;31:215-22. [Crossref] [PubMed]
- Chen YJ, Ko JY, Chen PJ, et al. Chromosomal aberrations in nasopharyngeal carcinoma analyzed by comparative genomic hybridization. Genes Chromosomes Cancer 1999;25:169-75. [Crossref] [PubMed]
- Hui AB, Lo KW, Leung SF, et al. Detection of recurrent chromosomal gains and losses in primary nasopharyngeal carcinoma by comparative genomic hybridisation. Int J Cancer 1999;82:498-503. [Crossref] [PubMed]
- Fang Y, Guan X, Guo Y, et al. Analysis of genetic alterations in primary nasopharyngeal carcinoma by comparative genomic hybridization. Genes Chromosomes Cancer 2001;30:254-60. [Crossref] [PubMed]
- Chien G, Yuen PW, Kwong D, et al. Comparative genomic hybridization analysis of nasopharygeal carcinoma: consistent patterns of genetic aberrations and clinicopathological correlations. Cancer Genet Cytogenet 2001;126:63-7. [Crossref] [PubMed]
- Hui AB, Or YY, Takano H, et al. Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res 2005;65:8125-33. [Crossref] [PubMed]
- Fan CS, Wong N, Leung SF, et al. Frequent c-myc and Int-2 overrepresentations in nasopharyngeal carcinoma. Hum Pathol 2000;31:169-78. [Crossref] [PubMed]
- Or YY, Hui AB, To KF, et al. PIK3CA mutations in nasopharyngeal carcinoma. Int J Cancer 2006;118:1065-7. [Crossref] [PubMed]
- Or YY, Chung GT, To KF, et al. Identification of a novel 12p13.3 amplicon in nasopharyngeal carcinoma. J Pathol 2010;220:97-107. [Crossref] [PubMed]
- Hui AB, Lo KW, Teo PM, et al. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol 2002;20:467-73. [PubMed]
- Chan AS, To KF, Lo KW, et al. Frequent chromosome 9p losses in histologically normal nasopharyngeal epithelia from southern Chinese. Int J Cancer 2002;102:300-3. [Crossref] [PubMed]
- Chan AS, To KF, Lo KW, et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from southern Chinese. Cancer Res 2000;60:5365-70. [PubMed]
- Lo KW, Teo PM, Hui AB, et al. High resolution allelotype of microdissected primary nasopharyngeal carcinoma. Cancer Res 2000;60:3348-53. [PubMed]
- Cheung AK, Lung HL, Ko JM, et al. Chromosome 14 transfer and functional studies identify a candidate tumor suppressor gene, mirror image polydactyly 1, in nasopharyngeal carcinoma. Proc Natl Acad Sci U S A 2009;106:14478-83. [Crossref] [PubMed]
- Lung HL, Lo CC, Wong CC, et al. Identification of tumor suppressive activity by irradiation microcell-mediated chromosome transfer and involvement of alpha B-crystallin in nasopharyngeal carcinoma. Int J Cancer 2008;122:1288-96. [Crossref] [PubMed]
- Cheng Y, Lung HL, Wong PS, et al. Chromosome 13q12 region critical for the viability and growth of nasopharyngeal carcinoma hybrids. Int J Cancer 2004;109:357-62. [Crossref] [PubMed]
- Cheng Y, Ko JM, Lung HL, et al. Monochromosome transfer provides functional evidence for growth-suppressive genes on chromosome 14 in nasopharyngeal carcinoma. Genes Chromosomes Cancer 2003;37:359-68. [Crossref] [PubMed]
- Cheng Y, Chakrabarti R, Garcia-Barcelo M, et al. Mapping of nasopharyngeal carcinoma tumor-suppressive activity to a 1.8-megabase region of chromosome band 11q13. Genes Chromosomes Cancer 2002;34:97-103. [Crossref] [PubMed]
- Cheng Y, Stanbridge EJ, Kong H, et al. A functional investigation of tumor suppressor gene activities in a nasopharyngeal carcinoma cell line HONE1 using a monochromosome transfer approach. Genes Chromosomes Cancer 2000;28:82-91. [Crossref] [PubMed]
- Cheng Y, Poulos NE, Lung ML, et al. Functional evidence for a nasopharyngeal carcinoma tumor suppressor gene that maps at chromosome 3p21.3. Proc Natl Acad Sci U S A 1998;95:3042-7. [Crossref] [PubMed]
- Yau WL, Lung HL, Zabarovsky ER, et al. Functional studies of the chromosome 3p21.3 candidate tumor suppressor gene BLU/ZMYND10 in nasopharyngeal carcinoma. Int J Cancer 2006;119:2821-6. [Crossref] [PubMed]
- Cheung AK, Lung HL, Hung SC, et al. Functional analysis of a cell cycle-associated, tumor-suppressive gene, protein tyrosine phosphatase receptor type G, in nasopharyngeal carcinoma. Cancer Res 2008;68:8137-45. [Crossref] [PubMed]
- Law EW, Cheung AK, Kashuba VI, et al. Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma. Oncogene 2012;31:728-38. [Crossref] [PubMed]
- Lo PH, Xie D, Chan KC, et al. Reduced expression of RASSF1A in esophageal and nasopharyngeal carcinomas significantly correlates with tumor stage. Cancer Lett 2007;257:199-205. [Crossref] [PubMed]
- Lung HL, Lo PH, Xie D, et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int J Cancer 2008;123:401-8. [Crossref] [PubMed]
- Cheung AK, Ko JM, Lung HL, et al. Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc Natl Acad Sci U S A 2011;108:8390-5. [Crossref] [PubMed]
- Chen H, Ko JM, Wong VC, et al. LTBP-2 confers pleiotropic suppression and promotes dormancy in a growth factor permissive microenvironment in nasopharyngeal carcinoma. Cancer Lett 2012;325:89-98. [Crossref] [PubMed]
- Kan R, Shuen WH, Lung HL, et al. NF-κB p65 Subunit Is Modulated by Latent Transforming Growth Factor-β Binding Protein 2 (LTBP2) in Nasopharyngeal Carcinoma HONE1 and HK1 Cells. PLoS One 2015;10:e0127239. [Crossref] [PubMed]
- Lung HL, Cheng Y, Kumaran MK, et al. Fine mapping of the 11q22-23 tumor suppressive region and involvement of TSLC1 in nasopharyngeal carcinoma. Int J Cancer 2004;112:628-35. [Crossref] [PubMed]
- Lung HL, Bangarusamy DK, Xie D, et al. THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene 2005;24:6525-32. [PubMed]
- Huang DP, Lo KW, van Hasselt CA, et al. A region of homozygous deletion on chromosome 9p21-22 in primary nasopharyngeal carcinoma. Cancer Res 1994;54:4003-6. [PubMed]
- Chung GT, Lung RW, Hui AB, et al. Identification of a recurrent transforming UBR5-ZNF423 fusion gene in EBV-associated nasopharyngeal carcinoma. J Pathol 2013;231:158-67. [Crossref] [PubMed]
- Valouev A, Weng Z, Sweeney RT, et al. Discovery of recurrent structural variants in nasopharyngeal carcinoma. Genome Res 2014;24:300-9. [Crossref] [PubMed]
- Chan KC, Ko JM, Lung HL, et al. Catalytic activity of Matrix metalloproteinase-19 is essential for tumor suppressor and anti-angiogenic activities in nasopharyngeal carcinoma. Int J Cancer 2011;129:1826-37. [Crossref] [PubMed]
- Wong VC, Chen H, Ko JM, et al. Tumor suppressor dual-specificity phosphatase 6 (DUSP6) impairs cell invasion and epithelial-mesenchymal transition (EMT)-associated phenotype. Int J Cancer 2012;130:83-95. [Crossref] [PubMed]
- Lung HL, Cheung AK, Ko JM, et al. Deciphering the molecular genetic basis of NPC through functional approaches. Semin Cancer Biol 2012;22:87-95. [Crossref] [PubMed]
- Li L, Zhang Y, Fan Y, et al. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics 2015;7:155-73. [Crossref] [PubMed]
- Wong TS, Tang KC, Kwong DL, et al. Differential gene methylation in undifferentiated nasopharyngeal carcinoma. Int J Oncol 2003;22:869-74. [PubMed]
- Ko JY, Lee TC, Hsiao CF, et al. Definition of three minimal deleted regions by comprehensive allelotyping and mutational screening of FHIT,p16(INK4A), and p19(ARF) genes in nasopharyngeal carcinoma. Cancer 2002;94:1987-96. [Crossref] [PubMed]
- Qiu GH, Tan LK, Loh KS, et al. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene 2004;23:4793-806. [Crossref] [PubMed]
- Ayadi W, Karray-Hakim H, Khabir A, et al. Aberrant methylation of p16, DLEC1, BLU and E-cadherin gene promoters in nasopharyngeal carcinoma biopsies from Tunisian patients. Anticancer Res 2008;28:2161-7. [PubMed]
- Liu XQ, Chen HK, Zhang XS, et al. Alterations of BLU, a candidate tumor suppressor gene on chromosome 3p21.3, in human nasopharyngeal carcinoma. Int J Cancer 2003;106:60-5. [Crossref] [PubMed]
- Lo KW, Kwong J, Hui AB, et al. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res 2001;61:3877-81. [PubMed]
- Jiang W, Liu N, Chen XZ, et al. Genome-Wide Identification of a Methylation Gene Panel as a Prognostic Biomarker in Nasopharyngeal Carcinoma. Mol Cancer Ther 2015;14:2864-73. [Crossref] [PubMed]
- Yang X, Dai W, Kwong DL, et al. Epigenetic markers for noninvasive early detection of nasopharyngeal carcinoma by methylation-sensitive high resolution melting. Int J Cancer 2015;136:E127-35. [Crossref] [PubMed]
- Tian F, Yip SP, Kwong DL, et al. Promoter hypermethylation of tumor suppressor genes in serum as potential biomarker for the diagnosis of nasopharyngeal carcinoma. Cancer Epidemiol 2013;37:708-13. [Crossref] [PubMed]
- Zhang Z, Sun D, Hutajulu SH, et al. Development of a non-invasive method, multiplex methylation specific PCR (MMSP), for early diagnosis of nasopharyngeal carcinoma. PLoS One 2012;7:e45908. [Crossref] [PubMed]
- Kwong J, Lo KW, To KF, et al. Promoter hypermethylation of multiple genes in nasopharyngeal carcinoma. Clin Cancer Res 2002;8:131-7. [PubMed]
- Zhou L, Jiang W, Ren C, et al. Frequent hypermethylation of RASSF1A and TSLC1, and high viral load of Epstein-Barr Virus DNA in nasopharyngeal carcinoma and matched tumor-adjacent tissues. Neoplasia 2005;7:809-15. [Crossref] [PubMed]
- Hutajulu SH, Indrasari SR, Indrawati LP, et al. Epigenetic markers for early detection of nasopharyngeal carcinoma in a high risk population. Mol Cancer 2011;10:48. [Crossref] [PubMed]
- Wang T, Liu H, Chen Y, et al. Methylation associated inactivation of RASSF1A and its synergistic effect with activated K-Ras in nasopharyngeal carcinoma. J Exp Clin Cancer Res 2009;28:160. [Crossref] [PubMed]
- Loyo M, Brait M, Kim MS, et al. A survey of methylated candidate tumor suppressor genes in nasopharyngeal carcinoma. Int J Cancer 2011;128:1393-403. [Crossref] [PubMed]
- Zhou W, Feng X, Li H, et al. Inactivation of LARS2, located at the commonly deleted region 3p21.3, by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Acta Biochim Biophys Sin (Shanghai) 2009;41:54-62. [Crossref] [PubMed]
- Kwong J, Lo KW, Chow LS, et al. Epigenetic silencing of cellular retinol-binding proteins in nasopharyngeal carcinoma. Neoplasia 2005;7:67-74. [Crossref] [PubMed]
- Li L, Tao Q, Jin H, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res 2010;16:2949-58. [Crossref] [PubMed]
- Ying J, Li H, Seng TJ, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene 2006;25:1070-80. [Crossref] [PubMed]
- Seng TJ, Low JS, Li H, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene 2007;26:934-44. [Crossref] [PubMed]
- Chang HW, Chan A, Kwong DL, et al. Evaluation of hypermethylated tumor suppressor genes as tumor markers in mouth and throat rinsing fluid, nasopharyngeal swab and peripheral blood of nasopharygeal carcinoma patient. Int J Cancer 2003;105:851-5. [Crossref] [PubMed]
- Tong JH, Tsang RK, Lo KW, et al. Quantitative Epstein-Barr virus DNA analysis and detection of gene promoter hypermethylation in nasopharyngeal (NP) brushing samples from patients with NP carcinoma. Clin Cancer Res 2002;8:2612-9. [PubMed]
- Wong TS, Kwong DL, Sham JS, et al. Quantitative plasma hypermethylated DNA markers of undifferentiated nasopharyngeal carcinoma. Clin Cancer Res 2004;10:2401-6. [Crossref] [PubMed]
- Wiest JS, Franklin WA, Otstot JT, et al. Identification of a novel region of homozygous deletion on chromosome 9p in squamous cell carcinoma of the lung: the location of a putative tumor suppressor gene. Cancer Res 1997;57:1-6. [PubMed]
- Lo KW, Cheung ST, Leung SF, et al. Hypermethylation of the p16 gene in nasopharyngeal carcinoma. Cancer Res 1996;56:2721-5. [PubMed]
- Chan SL, Cui Y, van Hasselt A, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest 2007;87:644-50. [Crossref] [PubMed]
- Cheung HW, Ching YP, Nicholls JM, et al. Epigenetic inactivation of CHFR in nasopharyngeal carcinoma through promoter methylation. Mol Carcinog 2005;43:237-45. [Crossref] [PubMed]
- Zhang X, Li W, Li H, et al. Genomic methylation profiling combined with gene expression microarray reveals the aberrant methylation mechanism involved in nasopharyngeal carcinoma taxol resistance. Anticancer Drugs 2012;23:856-64. [Crossref] [PubMed]
- Sengupta S, den Boon JA, Chen IH, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res 2006;66:7999-8006. [Crossref] [PubMed]
- Shi W, Bastianutto C, Li A, et al. Multiple dysregulated pathways in nasopharyngeal carcinoma revealed by gene expression profiling. Int J Cancer 2006;119:2467-75. [Crossref] [PubMed]
- Lo YM, Chan LY, Lo KW, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res 1999;59:1188-91. [PubMed]
- Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013;110:18761-8. [Crossref] [PubMed]
- Alajez NM, Shi W, Hui AB, et al. Enhancer of Zeste homolog 2 (EZH2) is overexpressed in recurrent nasopharyngeal carcinoma and is regulated by miR-26a, miR-101, and miR-98. Cell Death Dis 2010;1:e85. [Crossref] [PubMed]
- Song LB, Zeng MS, Liao WT, et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res 2006;66:6225-32. [Crossref] [PubMed]
- Tong ZT, Cai MY, Wang XG, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene 2012;31:583-94. [PubMed]
- Yan M, Zhang Y, He B, et al. IKKα restoration via EZH2 suppression induces nasopharyngeal carcinoma differentiation. Nat Commun 2014;5:3661. [Crossref] [PubMed]
- Zhong J, Min L, Huang H, et al. EZH2 regulates the expression of p16 in the nasopharyngeal cancer cells. Technol Cancer Res Treat 2013;12:269-74. [PubMed]
- Cai MY, Tong ZT, Zhu W, et al. H3K27me3 protein is a promising predictive biomarker of patients' survival and chemoradioresistance in human nasopharyngeal carcinoma. Mol Med 2011;17:1137-45. [PubMed]
- Tsai CL, Li HP, Lu YJ, et al. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res 2006;66:11668-76. [Crossref] [PubMed]
- Dutton A, Woodman CB, Chukwuma MB, et al. Bmi-1 is induced by the Epstein-Barr virus oncogene LMP1 and regulates the expression of viral target genes in Hodgkin lymphoma cells. Blood 2007;109:2597-603. [Crossref] [PubMed]
- Minarovits J. Epigenotypes of latent herpesvirus genomes. Curr Top Microbiol Immunol 2006;310:61-80. [Crossref] [PubMed]
- Matsusaka K, Kaneda A, Nagae G, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res 2011;71:7187-97. [Crossref] [PubMed]
- Kaneda A, Matsusaka K, Aburatani H, et al. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res 2012;72:3445-50. [Crossref] [PubMed]


