An overview of the NCI precision medicine trials—NCI MATCH and MPACT
Background
The concept of oncogene addiction was first proposed by Weinstein (1), and has led to a whole new approach to cancer treatment. The discovery of imatinib, the Bcr-Abl tyrosine kinase inhibitor, in the treatment of chronic myelogenous leukemia, revolutionized treatment paradigms with regard to targeted therapies, as this was the first targeted agent to illustrate the concept that treating the principal driving oncogene can have a powerful impact on response (2). More recent efforts to catalog driver mutations across the entire cancer population have led to the development of a plethora of targeted agents. Subsequent generations of molecularly targeted agents have effectively subcategorized tumors into smaller molecular subsets, such as EGFR and ALK inhibitors in non-small cell lung cancer and BRAF inhibitors in melanoma, in an effort to duplicate this success. As a further example, trastuzumab has received approval for gastro-esophageal and gastric cancers in addition to HER2 overexpressing breast cancers (3,4). These efforts have led to the realization that the targeting of these mutations has the potential to transcend tumor histologies, effectively categorizing tumors based on the molecular signature.
Recent advances in biotechnology and bioinformatics over the past decade have led to a greater appreciation for the heterogeneity of tumors and the complex signaling pathways involved in the resistance to treatment. This complexity requires a network-based streamlined approach to the interpretation of data generated from a profile of the tumor. The current challenge of clinical trial design is focused upon the identification of molecular alterations in tumors and the selection for those patients who would be most likely to benefit from a particular targeted therapy. The Division of Cancer Diagnosis and Treatment of the United States National Cancer Institute (NCI) has accepted this challenge and is presently engaged in several trials dedicated to precision-based medicine (http://dctd.cancer.gov/MajorInitiatives/NCI-sponsored_trials_in_precision_medicine.htm). The NCI Molecular Analysis for Therapy Choice (MATCH), Molecular Profiling-based Assignment of Cancer Therapy (MPACT), and Exceptional Responders study are among these trials.
NCI MATCH trial
The NCI MATCH trial was initiated as a broad-based genomic pre-screening study to assign patients whose tumors harbor specific molecular aberrations to relevant targeted treatments, without regards to tumor histology type. This trial aims to establish whether patients with tumor mutations, amplifications or translocations of interest are likely to derive clinical benefit if treated with agents targeting that specific molecular change in a one stage single-arm design. To provide the greatest opportunity to patients, this trial will cover a large range of mutations with matching options. In order to design such a complex trial, a panel of experts in developmental therapeutics, clinical trial design, genetic sequencing, molecular oncology, informatics, and statistics were consulted to develop an algorithm that would define clinical action based on genetic variants reported in the genes of interest. The structure of the study involves a master protocol to ensure the common elements of the subprotocols remain consistent across the arms. This study is additionally designed with the flexibility to open and close arms under the umbrella of the master protocol, with each arm treated as a separate phase 2 trial.
To ensure for adequate patient enrollment, the trial will be run through the NCI National Clinical Trials Network (NCTN) and NCI Community Oncology Research Program (NCORP). NCORP will help bring this nationwide study to patients treated in the community setting and increase accessibility to patients. The ECOG-ACRIN group will coordinate the trial for the NCTN, with broad representation through having separate principal investigators for each of the sub-protocols, each representing the different groups within the NCTN. The large portfolio of agents needed for the success of this trial required the participation of a multitude of pharmaceutical partners. The Cancer Therapy Evaluation Program (CTEP) of the NCI assisted in the coordination and contracting of these agents. The NCI Center for Biomedical Informatics and Information Technology (CBIIT) along with members of the NCI MATCH team generated the informatics structure for this trial. Multiple committees, including Agents and Genes Working Groups, Sample and Sequencing Network Working Group, and Protocol Logistics Working Group, among others, were established to concurrently develop the multitude of components for this massive endeavor.
NCI MATCH will accrue patients with solid tumors, with disease that has progressed following at least one line of standard systemic therapy, or for whom no standard therapy exists. As this is an exploratory trial, histologies for which there is already an FDA approved indication with that agent, or that have been shown to not respond to a particular agent, will accordingly be excluded from the corresponding agent. The study is designed to assign targeted treatment based on a biopsy obtained after enrollment. Molecular changes will be the selection criterion for entry to a particular arm. The study drugs included in this trial include single agents and combinations that have either received FDA approval or are investigational agents that have achieved at least a recommended phase 2 dose.
The NCI MATCH trial will collect somatic (tumor) genomic data from all patients enrolled through a screening biopsy. As tumors sometimes accumulate additional mutations after various treatments or with continued growth and metastasis, a biopsy closest to the time of initiating treatment will be pursued in order to obtain the most reflective state of the tumor. A biopsy after progression will also be pursued, with special interest in those patients who initially responded to treatment, to assist in understanding the mechanism of resistance. The Molecular Characterization (MoCha) Laboratory of the NCI was charged with development of the assay to identify these actionable mutations. The patient’s tumor biopsy will be screened for pre-defined variations in genes within a NCI MATCH CLIA-certified laboratory. The molecular profiling assays will include large-scale parallel tumor sequencing (next generation sequencing) strategies, including a targeted Ampliseq panel as well as other molecular assays such as immunohistochemistry (IHC). The selection of treatment will be rule-based and will be applied by a rigorously validated informatics system to derive a tentative treatment assignment. If a patient is ineligible for the original assigned treatment arm because of a pre-defined clinical ineligibility criterion, and patient’s tumor harbors additional abnormalities for which treatments are available on the study, the system algorithm will continue to provide assignments until all available options are exhausted (Figure 1).
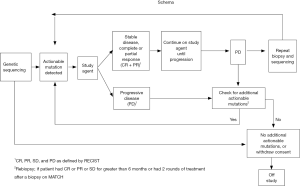
On this trial only malignant tissue will be screened. As such, definitive abnormalities in germline tissues (heritable diseases) cannot be identified with any certainty. Due to the concern that some of the genes tested may be of germline origin, a committee of multidisciplinary experts (genetics, oncology, bioethicists, patient advocates) was formed to address this ethical concern. Currently, findings will be communicated to the treating clinician with the recommendation to consider germline testing if clinical and/or family history is consistent with the presence of such an inheritable germline mutation. In many cases the medical significance of genetic variants are unknown (6,7). With the changing field of genomics, a steering committee has been tasked with monitoring the changing landscape.
This study affords a unique opportunity to collect information about the prevalence of mutations, translocations and amplifications in genes associated with cancer, and how these tumors respond to targeted therapy in the treatment-refractory tumor setting. DNA variants and changes in RNA expression from tumors collected at the point of progression on treatment is anticipated to illuminate resistance mechanisms that will inform subsequent studies and improve upon patient outcomes. NCI MATCH has opened in August 2015.
MPACT trial
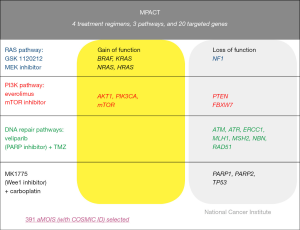
MPACT was designed to address the question of whether targeting an oncogenic driver would be more efficacious than not targeting the mutation. This pilot trial aims to establish whether advanced cancer patients who have exhausted all standard treatment options with proven benefit and have tumor harboring mutations in one of three main genetic pathways (DNA repair, PI3K, or RAS/RAF/MEK) are more likely to derive clinical benefit if treated with agents targeting that pathway than if treated with agents targeting one of the other pathways not identified to be dysregulated within the tumor. The agents administered in this trial are at recommended phase 2 dosing schedules. Currently the trial involves three pathways and four treatment arms (Figure 2): (I) veliparib (PARP inhibitor) with temozolomide for defects in the DNA repair pathway; (II) AZD-1775 (Wee1 inhibitor) plus carboplatin for defects in DNA repair pathway; (III) everolimus (mTOR inhibitor) for mutations in the PI3K pathway; or (IV) trametinib DMSO (MEK inhibitor) for mutations in the RAS/RAF/MEK pathway. Because of known benefits of BRAF inhibitor in melanoma and PARP inhibitors in BRCA ovarian cancer patients, these selected exclusions were built into the trial. The patients may remain eligible to be screened but will only be eligible to receive any of the study treatments if they have other actionable mutations.
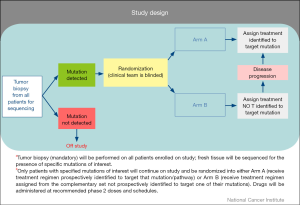
Similar to NCI MATCH, patients undergo tumor biopsies at the time of enrollment with the tumor sequenced in a CLIA-certified lab for actionable mutations. Distinct from NCI MATCH, patients for whom an actionable mutation is detected undergo a 2:1 randomization to one of two arms based on results of molecular profiling analysis (Figure 3) where the investigator and patients are blinded to the molecular target. Patients randomized to the treatment arm would receive drug or drug combinations designed to target the identified genetic mutation. Patients randomized to Arm B would receive drug or drug combinations not prospectively identified to target the identified mutation. Patients in whom no actionable mutations are identified in one of the three pathways (DNA repair, PI3K, or RAS/RAF/MEK) would be deemed ineligible for further treatment. Patients who have been treated and subsequently progress on their respective treatment arm will have their molecular profiling analysis unblinded and are permitted to crossover to the treatment arm if originally assigned to the control arm. Similar to NCI MATCH, emphasis is placed on repeat biopsy at time of progression to further understand the resistance mechanisms and whether exposure to targeted agents may have created a selection pressure for the acquisition of new lesions. Given the relative frequencies of mutations in the pathways of interest in this study, approximately 700 patients will be enrolled to acquire 180 evaluable patients with the initial four arms, assuming the population screened is similar between the treatment arm and the control arm. This trial is also designed to have flexibility with regard to the addition of new pathways/treatment arms. The endpoint of the study will compare the response rate [complete response (CR) + partial response (PR)] and/or 4-month progression-free-survival of the treatment arm versus the control arm. MPACT is currently open in the Developmental Therapeutics Clinic, NCI but will be available at other sites through the NCI-sponsored Experimental Therapeutics Clinical Trials Network (ETCTN) in the near future.
The backbone of both these precision-based medicine trials is heavily dependent upon having an accurate, reliable, and rapid molecular assay for the identification of actionable mutations. For MPACT, genetic sequencing will be performed in the CLIA-certified MoCha at the Frederick National Laboratory for Cancer Research (FNLCR). The genetic variants to be assessed and treatment algorithms have been prospectively defined to allow for assignment of specific treatment arms on study. For MPACT, 20 genes were selected for the initial analysis panel based on several criteria: (I) the biological pathway(s) affected by the targeted therapy were examined (pathways: RAS/RAF/MEK signaling pathway, PI3K/AKT pathway and DNA repair pathways; (II) genes within these pathways were selected based on demonstrating a minimum frequency (5%) of somatic variants as listed in the Catalogue of Somatic Mutations in Cancer (COSMIC) database; (III) genes known to modulate the targets of the study drugs; (IV) a Molecular Tumor Board review of the preclinical and clinical literature for the selection. A variety of specimen and assay quality checks are built into the assay process.
As the selection of treatment arm is rule-based, an informatics system, called GeneMed was designed to streamline the annotation of sequencing data, facilitate the review of variant mutations, and aid the identification of the actionable mutation. The results from the assay are processed based on predefined rules, and a treatment selection is assigned. If patient has two actionable mutations, the decision of which mutation will determine treatment selection is also rule-based.
Exceptional Responder
Despite best efforts, the majority of single agent anti-neoplastic drugs that enter phase 1 and 2 clinical trials ultimately fail to demonstrate sufficient efficacy to support further development. In rare exceptions, however, one or two patients achieve a significant response to the therapy or derive unexpected long-term benefit on these trials. These small subsets of responders from these otherwise “failed” trials/treatments may hold the key to insight in tumor biology and the identification of the molecular markers that predict for response to treatment. In support of this approach, several case reports have highlighted these “Exceptional Responders”. As an example, a urothelial cancer patient with a TSC1 and NF2 mutation achieved a durable CR to everolimus in a phase 2 trial that had failed to meet its phase 2 endpoint (8). Alterations in these genes were known to be associated with mTORC dependence in preclinical studies. The authors sequenced 13 additional patients with bladder cancer who had received everolimus, and found that 4 of 5 patients with TSC1 mutations had tumor shrinkage, whereas those without the mutation did not. A second patient with urothelial cancer, identified to have a novel mTOR mutation by whole exome sequencing, also had a CR to the combination of everolimus and pazopanib (9). These reports, as well as others in the literature, suggest that a search for elusive molecular targets in responders as a means to enrich studies for those patients most likely to benefit from any particular treatment holds promise for a more successful drug trial. The ability to identify molecular markers that are able to predict a clinical response in any particular subsets of patients will provide the tools necessary to conduct further studies consistent with the principles of precision medicine and allow for more rapid development of novel strategies.
The Exceptional Responders initiative aims to establish a repository of information on tumor biology based on data collected from these unique responders. The success of this endeavor depends upon having accurate and reliable demographics, clinical history, and response data for patients who have been treated, adequate tissue for analysis, robust analytical techniques/platforms, and appropriate bioinformatics/biostatistical tools. The Exceptional Responders project will collect tissues from patients who fit the definition of Exceptional Responders (Table 1) and use whole exome sequencing, and/or targeted NGS assay deep sequencing for full genomic analysis of patient tumors. If sufficient material is available, further exploration with additional analyses [e.g., whole genome sequencing, messenger RNA (mRNA)-sequencing, micro RNA (miRNA) sequencing, promoter methylation analysis, single nucleotide polymorphism (SNP) analysis, etc] will be performed. All data will be de-identified and placed in a controlled-access database to serve as a repository of information to allow investigators to mine data and design and build clinical trials around this information, based on molecular features predictive of benefit to a particular drug or drug class.

Full table
Factors to consider
The essential factors for the success of these NCI precision-based medicine trials bring to light fundamental issues inherent in transformative clinical trial design. The data generated from these trials have to be interpreted with certain assumptions: (I) target engagement by the selected agent has been confirmed either preclinically or clinically—even in best of circumstances, errors have been made especially in early development. One glaring example is that of iniparib, which failed to inhibit poly (ADP-ribose) polymerase in vitro, though this was not discovered until it had gone through phase 3 clinical development (10); (II) the assay used has been validated, confirmed to be reliable for the target, and transferable across sites—guidelines and standard operating procedures must be in place to ensure that biospecimen collection, storage, and processing meet quality standards for further sequencing and that regardless of where the specimen is handled, the same result can be expected. As an example, the lack of these institutional standards delayed initiation of the Cancer Genome Atlas initiative (11); (III) biopsy material obtained is representative of the entire tumor and metastatic site(s)—while driver mutations likely represent a significant portion of the existing tumor, tumor heterogeneity is a well-known challenge in the design of molecular targeted clinical trials. Even strategies such as serial tumor biopsies cannot completely eliminate this as a factor in the interpretation of data. Future development of circulating tumor cells or circulating DNA may help to further delineate driver mutations from those of bystander mutations. Radiographic record of biopsy sites may improve understanding of tumor heterogeneity; (IV) there is an available therapy for the target of interest—selection of the most reasonable agent for a specific target can sometimes be limited due to factors such as drug availability, ease of administration, or proven ability to combine with established agents in a particular clinical setting. Limitations in therapeutic options can further be complicated in situations of variants of unknown significance where benefit has not yet been confirmed.
The results of both NCI MATCH and MPACT will be informative and provide opportunities for further investigation. Though NCI MATCH’s primary endpoint is response rate, it is exploratory in nature. With a mix of histologies, including those of rare tumors, any response could provide interesting leads. How these leads will be explored and confirmed is not currently established. The strength of MPACT is heavily dependent upon accurate selection of the driver mutation in order to focus exploration of a particular pathway. Both trials contain multiple arms with small number of patients designed not as definitive trials but more as exploratory trials in order to guide further exploration of both tumor and pathways.
Conclusions
Advances in biotechnology and bioinformatics over the past decade have allowed for molecular characterization of patient tumors, opening opportunities for the development of tailored therapeutics based on characterization of a patient’s tumor. The Precision Medicine Initiative is a priority for the NCI, and was recently noted during President Obama’s 2014 State of the Union address (12). Currently the NCI is sponsoring several trials strategized to test the benefit of targeted therapy. NCI MATCH, MPACT and Exceptional Responder initiative are among these trials. Others including lung MAP and ALCHEMIST will be discussed by others in this journal. The results of the Exceptional Responders initiative in particular will be central to the identification of molecular features of tumors that would predict for response to a particular drug or class of drugs. For many currently standard chemotherapy drugs or regimens, the exact mechanism of action may not be known, and thus Exceptional Responders to such regimens may provide critical new data. The information obtained from this trial will be made available to investigators in a database that can be shared, built upon, and further mined. The NCI MATCH trial will evaluate these targets and whether they behave similarly across histologic subtypes. The provocative MPACT trial further seeks to address the larger question of the importance of targeting “actionable mutations” with targeted agents, and whether this will translate into meaningful clinical benefit above that achieved by current treatments.
The premise of both the NCI MATCH and MPACT trials relies heavily on the precision and accuracy of molecular tumor characterization techniques to find the target. Predefined rules allow for the elimination of bias from the selection process and allow for rapid decision-making once molecular targets are identified. Both trials also require a reliable informatics system to process the results of the assay and output of treatment selection. From a patient aspect, this process must additionally be sufficiently rapid to provide meaningful treatment options for patients willing to undergo biopsy and remain untreated while awaiting results. Additionally, with the continuing explosion of genomic data being generated, this cannot be a static process. The structure of these trials allows for flexibility with these changing data, allowing for addition of new variants and targets, and the removal of ineffective ones. By building in biopsies at the point of progression, these trials will also allow for a broader understanding of resistance mechanisms invoked with exposure to subsequent therapies and the intricate interplay between these molecular pathways.
Additional factors which need to be considered in implementation of these transformative trials involve the management of reporting of genomic data. Reporting of incidental findings to patients requires forethought, especially in situations of mutations of unknown significance. Results from these trials will provide a strong structure to build new and better treatment options for oncology patients in the twenty-first century. As a consequence, oncologists will increasingly be called upon to deal with assisting patients to understand the vast amount of genetic data generated from these studies and how best to use the information to assist in management of their cancer. With the evolution of vast amounts of information and the identification of smaller and smaller subpopulations of patients who would benefit from any one particular therapy, the question of how these particular treatments will garner regulatory approval remains to be seen.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science 2002;297:63-4. [PubMed]
- Doroshow JH, Kummar S. Translational research in oncology--10 years of progress and future prospects. Nat Rev Clin Oncol 2014;11:649-62. [PubMed]
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [PubMed]
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783-92. [PubMed]
- (DCTD) NCI-DoCTaD. NCI sponsored trials in precision medicine. Available online: http://dctd.cancer.gov/MajorInitiatives/NCI-sponsored_trials_in_precision_medicine.htm, accessed on June 2015.
- Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: meeting the challenge one bin at a time. Genet Med 2011;13:499-504. [PubMed]
- Caulfield T, McGuire AL, Cho M, et al. Research ethics recommendations for whole-genome research: consensus statement. PLoS Biol 2008;6:e73. [PubMed]
- Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science 2012;338:221. [PubMed]
- Wagle N, Grabiner BC, Van Allen EM, et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov 2014;4:546-53. [PubMed]
- Patel AG, De Lorenzo SB, Flatten KS, et al. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res 2012;18:1655-62. [PubMed]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931-45. [PubMed]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [PubMed]


