
BReast CAncer (BRCA) gene mutations as an emerging biomarker for the treatment of gastrointestinal malignancies
BReast CAncer 1/2 (BRCA1/2) and DNA repair pathways
BRCA1 and BRCA2 are tumor suppressor genes. Discovered in the early 1990’s as BReast CAncer susceptibility genes (1,2), they have been increasingly under the spotlight since 2013 after a renowned Hollywood actor announced her story (Angelina Jolie, “My medical choice”, New York Times; 2013). Among the scientific community there has been a growing interest in BRCA mutations following the discovery and development of poly-ADP ribose polymerase (PARP) inhibitors (PARPis) (3-5). It has been known for several decades that mutations in the BRCA genes increase the risk of breast cancer, ovarian cancer and other malignancies. A recent study showed that the cumulative risk of breast cancer by age 80 in BRCA1 and BRCA2 mutation carriers was as high as 72% and 69% respectively (6). Both have an increased risk of esophageal, gastric, cholangiocarcinoma (CCA) and pancreatic cancer (PDAC), only BRCA1 mutation carriers have an increased risk for colorectal cancer (CRC) (7-10). In a Korean population stomach cancer was the most common cancer site among BRCA carriers after breast and ovarian cancer (11).
BRCA mutations can be inherited as a germline mutation in an autosomal dominant fashion or can be acquired as somatic mutations in the tumor. Multiple variants of BRCA mutations have been identified in the population. A majority of the pathogenic variants are protein truncating variants (frame shift or stop gain variants) which induces a loss of function (12). BRCA genes follow Knudson’s two hit hypothesis where the second hit in the wild type allele of a germline mutation carrier is accrued by loss of allele or infrequently as a somatic mutation or promoter hypermethylation (BRCA1 only) (13). Germline inherited mutations have been associated with a younger age at first cancer diagnosis along with a higher proportion of independent cancer diagnoses compared to patients with wild type in the germline (13).
The increased risk of cancer in BRCA-mutated genes has been conferred primarily related to their role in DNA damage repair (DDR). The collective mechanisms by which a cell deals with DNA damage acquired during replication or exogenously is termed the DDR pathway. DNA damage can lead to three main consequences: (I) initiate pathways required to repair the damaged DNA; (II) activate factors to cause cell cycle arrest to allow time for cells to repair themselves; (III) direct cells with irreparable damage towards apoptosis pathways (14).
There are five main types of DDR pathways identified, namely base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination (HR) and non-homologous end joining (NHEJ) (15). DNA damage can be due mismatch during replication, single-strand breaks (SSB) or double-strand breaks (DSB). Replication errors and mismatch errors are often repaired through MMR. SSB, which are among the most common form of damage, are repaired through pathways initiated by PARPis (16). DSB’s are more lethal to the cells and need immediate repair primarily by two main pathways. The slower error free homologous recombination repair (HRR) pathway which uses an existing chromatid as a template and a NHEJ pathway (17).
It is essential to understand the HR pathway to understand certain unique characteristics of BRCA deficient tumors for effective targeting. DSB in DNA are generally identified by the Mre11-Rad50-Nbs1 (MRN) complex. A series of events are initiated beginning with activation of kinases like ATM (ataxia telangiectasia mutated), ATR and DNA-dependent protein kinase (DNA-PK). These kinases cause phosphorylation of numerous proteins including BRCA1 which promotes DNA end resection by nucleases like EXO-1 to form a 3' tail. BRCA1 then activates BRCA2 genes which helps recombinase RAD51 locate and bind to the single strand DNA to form a presynaptic filament (18). These filaments, with the help of BRCA1, invade into the sister chromatid to form a displacement loop or D-Loop (heteroduplex formation). D loops serve as a template for replication followed by holiday junction resolution of the heteroduplex complex (19,20). Beyond HRR, BRCA genes also play an important role in replication fork stability. In the absence of BRCA genes meiotic recombination 11-like (MRE11) nuclease leads to degradation of the replication fork causing increased genomic instability (21). Thus, BRCA deficient tumors must rely on error prone NHEJ pathway leading to high levels of genomic instability in tumors ultimately leading to cell death. Thus, in the absence of HRR pathway, tumors with BRCA mutations have increased sensitivity to agents that cause DSB like platinum-based chemotherapy, ionizing radiation and PARPis (22-24).
PARP are enzymes that are involved in DNA repair pathways. These enzymes are activated when endogenous or exogenous factors cause SSB or DSB. PARP binds to SSB and leads to further binding of branched poly-ADP ribose (PAR) chains in a process called PARylation (17,20). PARPis work on the principle of synthetic lethality which is a concept defined as early as 1945 (25). Synthetic lethality essentially refers to a set of two genes or molecular pathway where damage to either gene or pathway is not lethal but when there is damage to both, cell death occurs (26). PARP enzymes and BRCA genes are such a pair which are essential for SSB and DSB repair respectively. In the absence of both components there is accumulation of SSB which are converted to DSB during S phase and later leading to replication fork collapse (27) followed by activation of apoptosis pathway. When PARP enzymes are attached to SSB, followed by exposure to PARPis, the phenomenon of “PARP trapping” occurs which is more toxic to cells than being deficient in PARP (28). PARP enzymes are also involved in activating DNA-PK which leads to activation of NHEJ, the alternate pathway involved in DSB repair in cells where HRR is interrupted (29). Collectively, these mechanisms underpin the rationale for the exquisite sensitivity observed in tumors with BRCA gene mutation exposed to PARPis (30).
BRCAness, homologous recombination defect (HRD)ness and PARPness
The term “BRCAness” was first coined by Turner et al. in 2004 to explain the features exhibited by cancers that were BRCA mutated and to include cancers that exhibited similar characteristics without a specific BRCA mutation (31). Their original paper “Hallmarks of BRCAness” reported subtle phenotypic characteristics seen in some of the BRCA mutated breast cancers like basal cell type, high histological grade, higher chance of being triple negative [estrogen receptor (ER), progesterone receptor (PR), HER-2 negative], presence of “pushing margins” showing an increased tendency towards lymphocytic infiltration and genotypic features with more TP53 mutations, amplification of c-MYC and lack of ERBB2 and clinically by sensitivity to DNA damage inducing agents like cisplatin and mitomycin C. The presence of similar defining features were identified in cancer cells without BRCA mutations but with epigenetic changes like de novo methylation (silencing) of BRCA1 promoter region, EMSY amplification and in cells with other defects in the same DNA repair pathways modulated by BRCA genes, defining a group of cancers with BRCAness with serious clinical implications like sensitivity to specific chemotherapy and prognosis (31).
Following the approval of the first PARPi (olaparib in ovary cancer) in 2014 initially by the European Medicines Agency (EMA) (32) followed by the Food and Drug Administration (FDA) (33), there has been a growing interest in the scientific community to further explore the function of BRCA1 and BRCA2 genes and their role in DDR via the homologous repair pathway and others. Advances in whole genome sequencing (WGS) have led to the development of HRDetect by Davies et al. using six specific mutational signatures which allows detection of BRCA deficient tumors with a 98.7% sensitivity to broaden the spectrum of BRCAness tumors (34). Pilié et al. has proposed the broader term HRDness (35) to include the group of cancers that are sensitive to PARPis in the absence of BRCA mutations or BRCA like phenotypes but also loss of function of other genes which are not canonical DDR genes, or presence of oncometabolites (36) and lead to a HRD. Studies have shown that DDR is affected more in cells exposed to PARPis than cells completely lacking PARP due to a phenomenon called “PARP trapping” (27,28). Cerrato et al. (37) described multiple factors that act as a surrogate for PARPi sensitivity like TMPRSS2: ERG (38), EWS fusions (39), CDK12 attenuation (40), low expression of ERCC1 (41) and BAP1 deficiency (42) to name a few. Pilié’s group also proposes another term “PARPness” to describe markers predicting PARP sensitivity which are not involved in the HRR pathway like expression of Schlafen 11 (SLFN11) (35) or E-cadherin, or NAD+ depletion (43).
BRCA in gastrointestinal (GI) cancers
Despite major advances in cancer detection and treatment, GI cancers continue to have one of the lowest 5-year survival rates according to the Centers for Disease Control and Prevention (CDC) data. PDAC followed by primary liver cancer [hepatocellular cancer (HCC)], CCA and esophageal cancer lead the order regarding lethality. There is a growing trend towards personalization of cancer therapy along with integration of targeted therapy and supportive care to maximize the quality of life of life and survival. Precision medicine in oncology refers to utilizing the molecular profiles and biologic characteristic of individual tumor to guide therapeutic choices. Modalities to detect specific driving mutation and target them are being refined and developed. Amongst GI cancers, PDAC continues to have least favorable outcome, nevertheless, understanding of DNA repair mechanisms is most advanced in this disease. This has translated into more studies in periclinal and clinical settings with recent translation into improved outcomes for subsets of patients.
PDAC
Risk of PDAC and BRCA
A year after the discovery of BRCA2 gene, Goggins et al. in 1996 analyzed BRCA germline mutations in PDAC patients and identified a prevalence of 7.3% of cases with BRCA2 (44). The Breast Cancer Linkage Consortium also reported a relative risk (RR) of 3.51 in patients harboring BRCA2 mutations (8). Later the cumulative age adjusted lifetime risk PDAC with BRCA1 mutations was reported to be 3.6% in 2002 (9). In an unselected population about 6–9% of the PDAC are associated with BRCA1/2 mutations (45). Waddell and colleagues performed WGS and copy number variation analysis on 100 samples and classified PDAC into 4 subtypes based on structural rearrangements (45). Stable, locally rearranged, scattered and unstable subtypes. In the unstable group constituting 14% of cases, the majority, about 70%, had a BRCA signature. Another study which looked at mutational signatures to classify PDAC, identified that about 10% of cases had HRD and were termed as the DSB group (46). The mean age of diagnosis of 62 in individuals with BRCA mutations was about 10 years lower than the age reported in SEER data for PDAC (47). A deep sequencing study in PDAC individuals identified ~4% patients with germline BRCA2 mutation and another 3.9% with somatic BRCA mutations (48). A more promising study from Kondo’s group had noted 47% of patients in a small cohort of 28 consecutive patients with PDAC to have HRR related using a next generation sequencing assay (23).
Therapy and BRCA in PDAC
In a retrospective analysis of a large cohort of patients with PDAC and a germline BRCA mutation, Golan et al. reported an overall survival (OS) benefit in patients treated with platinum-based regimen compared to others supporting the hypothesis of the predictive significance of BRCA (49). The superiority of platinum-based regimen in tumors with BRCAness has since been replicated in multiple studies (50). Although, in surgically resected BRCA associated PDAC no difference has been noted in OS compared to wild type. However, was a trend towards increased disease-free survival in patients who received platinum-based chemotherapy in the BRCA mutated group (51). Hence the prognostic significance of BRCA is not well defined.
The first reports demonstrating the effectiveness of PARPi in PDAC surfaced in 2011 (52). Kaufman et al. conducted a pivotal trial which led to the FDA approval of PARPi in ovary cancer. In this trial a cohort of PDAC patients with a germline BRCA mutation were treated with single-agent olaparib and a response rate of 22% was observed in a group of individuals that had already received an average of two lines of prior therapy (33). This led to series of clinical trials evaluating the efficacy of PARPis (53,54). Notably, Yarchoan et al. evaluated olaparib in combination with irinotecan, cisplatin and mitomycin C and reported an overall response rate (ORR) of 23% among evaluable patients (55). However, myelosuppression was significant and dose-limiting, and the combination was halted from further development. A concerning fact was that two out of the three patients who had received more than 12 cycles of therapy and had an objective response had developed myelodysplastic syndrome of which one progressed to acute myeloid leukemia and died about 5 years after the start of treatment for PDAC. Another notable study was a phase II trial by Lowery and colleagues in 16 patients with known germline BRCA1/2 or PALB2 with prior treatment who received single agent veliparib, but failed to show any overall improvement in survival; however, all but one of these patients were platinum exposed/resistant likely accounting for the low objective response rate (56). The authors had proposed multiple reasons for these findings including the likelihood of olaparib being a stronger PARPi and a higher proportion of people with platinum resistance included in this study. The RUCAPANC trial, a phase 2 study in germline or somatic BRCA mutated PDAC patients who had received prior lines of treatment showed a response rate of 16% and a disease control rate of 32% with single agent rucaparib (57). Major grade ≥3 adverse events noted where anemia in 32% and fatigue in 16% of the patients. Interestingly in an untreated population with advanced PDAC a phase I trial by O’Reilly et al. with veliparib, cisplatin and gemcitabine showed an objective response rate of 78% in the BRCA mutated cohort (58). The phase 2 of this randomized trial NCT01585805 evaluating cisplatin, gemcitabine with/without veliparib in front-line BRCA1/2 or PALB2 mutated pancreas cancer has since completed accrual and demonstrated a very high response rate in both treatment arms 74.1% and 65% (P=0.55) for the triplet vs. the doublet, median PFS of 10.1 vs. 9.7 months (P=0.73) and OS of 15.5 vs. 16.4 months (P=0.6) (59). The triplet of cisplatin, gemcitabine and veliparib incurred significantly more grade 3–4 hematologic toxicity. Of further note, the 2-year survival rate for the combined study cohort was 30.6% and 3-year OS of 17.8%. Collectively these data endorse the value of platinum-based therapy in BRCA/PALB2 mutated PDAC and endorse cisplatin/gemcitabine as a standard treatment option, and an alternative to mFOLFIRINOX, in this patient population.
It is key to note that in most of these studies responses where seen in patients who were platinum-sensitive rather than resistant or refractory leading to the question of the best timing for introduction of PARPis. The POLO (Pancreas cancer OLaparib Ongoing) provides some insight into this question. This randomized, double blind, placebo-controlled phase III trial in metastatic PDAC looked at olaparib in a maintenance setting for patients with a known germline BRCA mutation and without disease progression after 16 weeks or greater therapy with platinum-based agent as first line treatment and has shown promising results (60). One hundred and fifty-four patients across 12 countries were randomized as 3:2 in favor of olaparib vs. placebo. The primary endpoint of progression-free survival (PFS) was 7.4 months in the olaparib vs. 3.8 months in the placebo arm with a hazard ratio for disease progression or death of 0.53 with a 95% confidence interval (CI): 0.35–0.82 and P=0.004. The median duration of response of 24.9 months in the olaparib arm compared to 3.7 months with placebo is very notable in this disease. On an intent to treat analysis thus far no difference in OS has been identified for the olaparib-treated patients over placebo in an interim analysis at 46% data maturity. Albeit, some experts have commented regarding PFS as being an inadequate end point in maintenance therapy trials compared to OS (61). Another paper from the same study which analyzed the health-related quality of life using global health scale found that there was no statistical difference between the two groups during the first 6 months of treatment [between-group difference −2.47; 95% CI: −7.27 to 2.33; P=0.31] meaning that in the maintenance setting olaparib was able to achieve significant PFS while maintaining overall quality of life (62). One other important point of discussion related to the POLO trial pertains to the use of a control arm of placebo rather than continuation of cytotoxic therapy, which in many parts of the world is a standard approach for these patients. Of specific note in late 2019, the FDA approved olaparib as a maintenance therapy following 4 months of platinum-based treatment in germline BRCA-mutated pancreas cancer based on the results of the polo trial.
Current National Comprehensive Cancer Network (NCCN) guidelines endorse routine germline testing for all individuals diagnosed with PDAC based on recently published data (63-65). In 2019 implemented this recommendation for universal germline testing, a recommendation which is independent of age, ethnicity, heritage, or personal or family history of malignancy. In addition, somatic profiling is recommended for patients who are candidates for further treatment. Until recently germline testing was only recommended in patients of high-risk groups like Ashkenazi Jewish descent and individuals with personal history or a strong family history of breast, ovarian or PDAC in one or more family members. These recommendations came in the light of new evidence from studies that have demonstrated that a significant number of patients with genetic mutations and potentially targetable heritable mutations in PDAC were identified even in individuals without a strong family or personal history of cancer. In a study published in JAMA, Hu et al. identified 21 cancer predisposition genes by genomic sequencing from peripheral blood in 3,030 adults diagnosed with PDAC included in the mayo clinic registry spanning over 16 years (65). They identified 6 genes with significantly higher association with PDAC, including BRCA1 and BRCA2 in 5.5%. However, interestingly while these genes existed in about 7.9% of patients with a family history of PDAC about 5.2% of patients without such a history also had these genes uncovering a large group of individuals who would have been missed.
Pishvaian and colleagues analyzed data from Know Your Tumor program which collected tumor samples from 640 patients from 287 different centers (66). These tumor samples where send for next-generation sequencing (NGS) and immunohistochemistry (IHC) analysis revealing 27% of these samples with highly actionable targets. Of these 8.4% of the actionable targets where in DNA repair genes BRCA1/2 and ATM. In patients who received matched therapy significantly longer PFS was observed compared to those who received unmatched therapy; PFS 4.1 vs. 1.9 months (hazard ratio, 0.47; 95% CI: 0.24–0.94; Padj. =0.03), making a strong argument in favor of precision oncology. As part of the same initiative their group analyzed 820 patients with PDAC in whom comprehensive genetic testing data was available to clearly define the prognostic and predictive role of HRD in PDAC. They reported that no significant difference was seen in median OS in platinum naïve patients with HRD and without HRD both in resected disease and advanced disease (67). This meant that HRD did not carry a prognostic significance in the absence of platinum exposure. Even though no significant difference in OS was noted between the two groups that received platinum-based therapy in surgically resected patients, there existed a very significant difference in advanced disease between HRD and without. Among the 311 patients with advanced disease who received platinum-based therapy the median OS was 2.37 years for patients with HRD vs. 1.45 years for patients without defects in HRD or DDR pathway [P=0.000072; hazard ratio, 0.44 (95% CI: 0.29–0.66)]. This study has now very clearly established the predictive value of HRD in pancreas cancer and the fact that the survival benefit is lost in platinum naïve patients calling for genetic testing in all patients.
The 2019 American Society of Clinical Oncology (ASCO) meeting at Chicago witnessed a sudden boom in that 3 different works were presented on pancreas cancer in the realm of HRD. Using MSK-IMPACT data Park and colleagues prospectively followed 461 patients and analyzed their outcomes with regards to germline and somatic HRD status (68). Although the OS was not different among patients that received platinum vs. non platinum-based regimen in the first-line setting, the OS was found to be better among patients harboring a germline HRD vs. patients without the defect irrespective of first-line platinum-based therapy. In the updated analysis additional predictors of response to platinum therapy included biallelic status and the presence of a core gene mutation in BRCA1/2 or PALB2 (69). Like Pishvaian’s group the investigators identified that OS was also significantly higher in the germline or somatic HRD patients. Chiorean and colleagues presented data from their phase II trial in metastatic PDAC who received a combination of modified FOLFIRI with veliparib vs. FOLFIRI in the second-line setting (70). One hundred and eight of 123 patients analyzed as per protocol at 35% of PFS events did not reveal a superiority with the combination compared to the control arm. 9% of the analyzed patients had HRD and about 30% of all patients had a defect in DDR genes. Pishvaian’s group also had promising results from their phase I/II trial combining veliparib with 5-FU and oxaliplatin (mFOLFOX6) in metastatic PDAC in 64 patients (71). The study achieved its primary end point of ORR ≥25% and was well tolerated. Notably the ORR in platinum naïve patients with a family history of breast or ovarian cancers and or DDR gene defects was about 58%. The results on these completed trials have been summarized in Table 1.
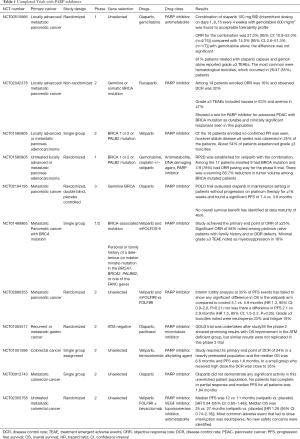
Full table
Summing up in PDAC, there are many ongoing trials further exploring DDR and treatment targeting possibilities. We anticipate that moving forward there will be an integration of both germline and somatic profiling information at the time of diagnosis to optimally define a treatment strategy for an individual patient.
CCA
Risk of CCA and BRCA and outcomes
The Breast Cancer Linkage Consortium had reported an increased incidence of CCA in BRCA2 mutated individuals with a RR of 4.97 (8), later confirmed by other studies (72). Mutations in the BRCA1 associated protein, BAP1, have been reported in intrahepatic cholangiocarcinoma (ICC) with a prevalence as high as 14.3% in several studies (73-76). In patients with ICC, low BAP1 expression has been associated with an aggressive biology, early recurrence post-surgery and poorer prognosis (77,78). In 2019, Lin and colleagues identified mutations in DDR genes in 26% of analyzed patients with primary liver cancers including HCC (80% of cases), ICC and hepatocellular CCA. In the ICC group BRCA1/2 mutations were detected in 9% of the individuals (79). About 6.7% of the 357 patients with primary liver cancer had at least one actionable target (79). This group of individuals with targetable mutations was notably higher at 47% in an analysis reported by Lowery and colleagues in CCA using the MSK-IMPACT platform (80). Parasramka et al. in 2017 were able to go a step further and show that ICC cell lines with low BAP1 expression was associated with increased sensitivity to gemcitabine and olaparib and uncovered the presence of a synergistic effect with the combination of the two (81). Multiple ongoing trials are underway which have been summarized in Table 2.
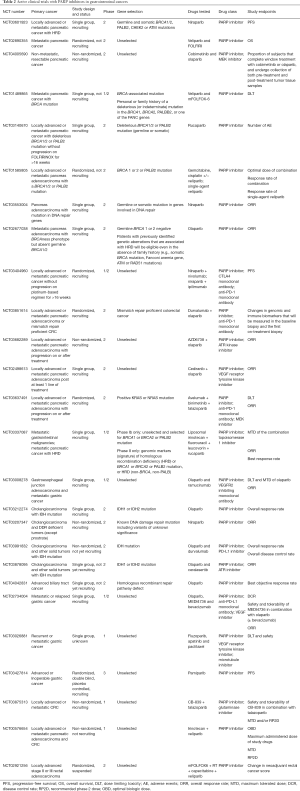
Full table
Therapy, BRCA and biliary cancers
Overall, there are relatively limited data regarding DDR-targeted approaches in biliary cancers. A multicenter retrospective cohort study in patients with germline or somatic mutations in BRCA genes showed superior outcomes in patients who received platinum-based regimens and/or PARPis with a median OS in stage I/II of 40.27 months and stages III/IV as 25 months (82). PARPis have also been shown to sensitize CCA to radiation therapy even in the absence of BRCA mutations (83). Targeting strategies in biliary cancers are early in development, however, this disease is recognized as one where platinum agents and PARPis may have a key role moving forward.
Esophageal cancer
Risk of esophageal cancer and BRCA
In 2007 germline DNA analysis of 197 Turkmen with esophageal squamous cell carcinoma (ESCC) identified BRCA2 mutations in 7.6% of cases (84). Later in 2011 Moran et al. published the first study which demonstrated an increased RR of esophageal cancer in BRCA1 mutation carriers. They reported a RR of 2.9 in BRCA1 carriers and a RR of 4.1 in BRCA2 carriers (85). Familial ESCC has also been found to have an increased frequency of BRCA2 mutations in a study from china (72). WGS analysis in esophageal adenocarcinoma showed three dominant categories based on mutational signatures, of which the DDR impaired cells lines comprised of 18% of the analyzed samples and showed promising results when treated with the PARPi olaparib combined with a DNA damaging agent, topotecan (86). Deng et al. in 2019 studied the germline mutational profile of 77 individuals with ESCC and found that 11.7% of patients had a HRD which was associated with a well differentiated tumor and a greater degree of lymph node metastasis (87). These studies have demonstrated a potentially actionable group for further exploration with PARPi. Miyamoto et al. in 2019 were able to demonstrate a synergistic effect of olaparib in ESCC cell lines when combined with chemo therapeutic agents (88).
Currently there is a relative dearth of trials in esophageal cancer although a clear actionable sub-group is evident in this disease type. We anticipate that further clinical data will ensue soon as interest in DDR genomic based treatment strategies expands in GI cancers.
Gastric cancer
Risk of gastric cancer and BRCA
The breast cancer linkage consortium study in 1999 reported an increased risk of gastric cancer in BRCA2 carriers with a RR of 2.59 while similar results were not observed with BRCA1 (8). Other studies have described BRCA1 as conferring an increased risk of gastric cancer (89,90). BRCA1 carriers were reported to have a cumulative age adjusted lifetime risk of 5.5%, four times the observed risk in the general population (9). BRCA1 nuclear expression was evaluated by Kim et al. in 2013 who found that decreased nuclear expression was associated with more advanced disease and perineural invasion and served as a marker for poor prognosis (91). They also observed that adjuvant chemotherapy was able to overcome this adverse prognosis and that decreased BRCA1 nuclear expression could serve as a predictive marker for response to adjuvant chemotherapy. Chen et al. identified that gastric cancer cells had BRCA1 protein detected in the cytoplasm as compared to nucleus and that decreased expression was associated with response to platinum-based therapy (92). Similarly, decreased expression of BRCA1 associated protein (BAP) was also found to correlate with advanced disease and poor prognosis and a potential prognostic marker (93). Recent evidence suggests that gastric cancer cells with lack of BRCA2 expression is associated with younger age and signet ring cell variant (94).
Alexandrov’s group analyzed gastric cancers for mutational signature 3, a base substitution signature that has been observed to be associated with BRCA mutations and serving as a surrogate for platinum sensitivity including in individuals without BRCA mutations (45). They noted that as many as 7–12% of gastric cancers had signature 3 which demonstrated the hallmarks of cancers with HRR in the absence of BRCA mutations and could potentially be targeted (95). Mihailidou’s group in 2017 suggested that gastric cancers with BRCA deficiency are susceptible to c-MET inhibition in the presence of DNA damaging agents (96). BRCA1 expression loss was associated with a poorer prognosis and had a 2-year survival rates close to 50% of that seen among BRCA1 wild type tumors (97). Using IHC Wang et al. showed that high BRCA1 and BRCA2 expression in cytoplasm predicted a favorable prognosis in gastric cancers while increased BRCA1 expression in the nucleus was shown to have a poor prognosis (98). In vitro analysis performed by Kim et al. showed an inverse correlation with BRCA1 expression level sensitivity to platinum agents (99).
Therapy, BRCA and gastric cancer
Borrowing ideas from their mantle cell lymphoma study Kubota et al. in 2014 demonstrated that gastric cancer cells lacking expression of ATM gene had increased sensitivity to olaparib (100). Bang et al. applied this in vivo and a phase II trial showed an increased OS with olaparib/paclitaxel combination in study population and not just the ATM low expression subgroup (101) paving way to a phase III trial, olaparib in combination with paclitaxel in patients with advanced gastric cancer who had progressed following first-line therapy (GOLD) trial. The GOLD trial was a double-blind, randomized, placebo-controlled phase III study which failed to demonstrate an improved OS with olaparib/paclitaxel vs. placebo/paclitaxel in the overall population [median OS 8.8 months in the olaparib group vs. 6.9 months in the placebo group; hazard ratio, 0.79 (97.5% CI: 0.63–1.00); P=0.026] and in the ATM deficient population [median OS 12.0 months in the olaparib group vs. 10.0 months in the placebo group; hazard ratio, 0.73 (97.5% CI: 0.40–1.34), P=0.25] (102). In vitro studies with combination of phosphoinositide 3-kinase (PI3K) and olaparib in cells with AT-rich interactive domain containing protein 1A (ARID1A) deficiency have shown promising results and would need further studies (103).
Like other GI cancers we anticipate an expansion of trials targeting DDR pathways in gastric cancer in the proximate future.
CRC
Risk of CRC and BRCA
The association between BRCA genes and CRC is controversial. Studies by Yurgelun et al. (104) had demonstrated an increased risk of CRC in BRCA1/2 mutation carriers while other studies like Brose et al. (9) demonstrated an increased risk only in BRCA1 mutation carriers. Phelan et al. (105) noted that the incidence of CRC was 4 times higher in women less than 50 years with mutated BRCA1, however no meaningful association was found in other groups (106,107). A comprehensive systematic review and metanalysis by Oh and colleagues showed no increased risk of CRC in BRCA2 mutation carriers and a 1.49-fold increased risk in BRCA1 mutation carriers (108). A more recent study demonstrated that 15% of the CRC analyzed had DDR gene alterations (109).
Preclinical and clinical data in CRC
Davidson et al. in 2013 noted that PARPis acted synergistically with platinum agents in vitro conditions (110) followed by Shelton et al. who showed in vivo response as well as an increased sensitivity to radiation therapy in the presence of PARPis (111). A preclinical study showed increased sensitivity of CRC cell lines with ATM deficiency to PARPi (112). However, a phase 2 trial in an unselected population failed to show any substantial activity with a single agent PARPi regardless of the microsatellite status (113). In a heavily pretreated population Wang and colleagues in their phase 2 trial achieved a disease control rate of 24% using a combination of veliparib with temozolomide (114). In a randomized, blinded, placebo-controlled phase 2 trial by Gorbunova’s group the addition of veliparib to first-line standard of care FOLFIRI ± bevacizumab in metastatic CRC did not reveal an improvement in PFS; 12 vs. 11 months (veliparib vs. placebo) [hazard ratio, 0.94 (95% CI: 0.60–1.48)] (115). The authors concluded that the increased incidence of hematological toxicities due to addition of the PARPi with chemotherapy had resulted in a shorter treatment duration.
Moving forward the value of DDR targeted approaches in CRC remains unclear. Arguably a population of interest is the subgroup with microsatellite instability from either a germline or somatic etiology and may represent a subgroup where such targeted approaches are relevant along with the small subset of patients harboring a germline BRCA mutation where CRC arises.
Looking forward
During the last few years of research, it has become more evident that not all individuals with BRCA mutations have the same response to PARPis or DNA damaging agents. Lord et al. proposed that there is a growing need for functional biomarkers for BRCAness given the differential sensitivity seen with PARPis in patients with hypermethylation of promoter region of BRCA in comparison to patients with germline mutations (116). This is in part because BRCA mutation status is not synonymous with HRD. Curtin and colleagues explain this in their paper on why BRCA mutation is not a tumor agnostic biomarker (117). This would explain why many recent trials did not produce the expected results or failed to show an efficacy or OS benefit (56,102,115). An ideal biomarker would be one which detects the presence of errors in DDR pathway and correlates well with PARPi sensitivity (118). Another concern is the development of second reactivating or reversion mutations during treatment (119), hence there needs to more studies on strategies to overcome resistance or better prevent or retard the development of resistance. IHC detection of RAD51 foci was a method proposed and evaluated in multiple studies. A major drawback is that RAD51 is not necessarily expressed in the nuclear foci of normal cells until there is DNA damage. Mutational signature 3 has been well correlated with HRD and might hold the key for population selection than merely the presence of germline or somatic BRCA mutation. Johnson et al. in an important recent paper, has postulated that not all pathogenic BRCA germline mutations are drivers of cancers and may be mere passengers based on their observation that as many as 8% of the cancer cells had lost the pathogenic germline BRCA mutation due to somatic loss of allele mutations (13). They also noted that only four types of cancers were enriched in germline BRCA mutation carriers after ancestry-adjusted association was applied namely breast, ovarian, prostrate and PDAC which they named BRCA associated cancers. They used HRD scores generated from mutational signatures or large-scale transitions in copy number alterations to evaluate different tumors. This led to an important hypothesis from the analysis that there was a near complete absence of HRD in non BRCA associated cancers (cancers beyond the four types afore mentioned) with heterozygous BRCA mutation that were not the cause of tumorigenesis but often produced somatically during tumor evolution. Research is underway to identify new molecules that act as surrogate marker for PARPi sensitivity and the presence of recombination deficiency.
Another area of growing interest has been the use of combination strategies with PARPis including with VEGF inhibitors and immunotherapy. The proof of concept studies for combination of PARP with VEGF inhibitors comes partly from the preclinical studies which demonstrated that hypoxia causes cells to develop HRD (120,121). In ovarian cancers, Liu and colleagues were able to demonstrate this clinically in a phase 2 study that showed significantly longer PFS (17.7 vs. 9.0 months, hazard ratio, 0.42; P=0.005) (122). Multiple clinical trials are ongoing to explore this concept (NCT02498613, NCT03008278, NCT03026881). Combining DNA repair inhibitors with Immunotherapy is an emerging field (123-126). In DDR deficient breast cancer, Parkes et al. identified a novel mechanism by which cytosolic DNA leads to activation of STING innate immune response upregulating the PD-L1 expression and making an argument for immune check point inhibitors in this subset in combination with S phase DNA damaging agents (123). In the field of urothelial cancer, Teo and colleagues identified a strong association between DDR alterations and response to anti-PD-L1 with a higher ORR in DDR deficient subtype compared to those without any DDR alterations (67.9% vs. 18.8%; P=0.001) (124). Mutations in the RAS signaling pathway was another suggested mechanism of PARPi resistance leading to combination with MEK inhibitors (NCT04005690, NCT03637491). Inspired from preclinical data, newer trials are combining PARPi with durvalumab, nivolumab and ipilimumab to test this hypothesis (NCT03851614, NCT03991832, NCT03404960).
Other ways of targeting BRCAness beyond PARPis are under investigation. RAD52 inhibitors have been found to be synthetically lethal in cells with BRCA1, BRCA2 and PALB2 mutations (127-130). WEE1 is a tyrosine kinase inhibitor of CDK1/2 which leads to arrest of mitosis in cells that sustain DNA damage. WEE1 inhibitors as monotherapy and in combination with ATR inhibitors have rendered cells sensitive to PARP inhibition and platinum agents (131). In PDAC a combination of a WEE1 and PARPi acts as a radiosensitizer (132). BRCA deficient cells switch to the alternative error prone pathway, NHEJ, and inhibition of polymerase theta has been shown to inhibit NHEJ and serve as a potential target (133). Inhibitors against DDR proteins like MTH1 and DNA damage signaling inhibitors like ATR, ATM, CHK1, USP7 (134) are all being studied (135). Table 3 provides a summary of relevant ongoing trials.
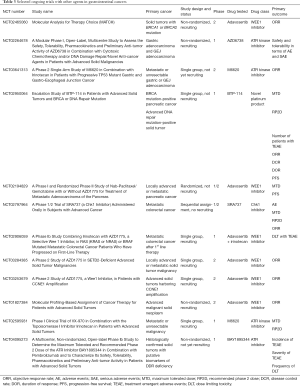
Full table
There have been major developments in the last couple of years regarding the recognition of BRCA mutations, their association with GI cancers and the targeted therapy implications. Importantly, a proof of principle has been secured with the recent FDA approval for olaparib in the setting of BRCA-related pancreas cancer, and even prior to that routine guideline endorsement for universal germline (and somatic) profiling in that disease. More routine evaluation of HR gene mutations across the spectrum of GI malignancies is warranted, including more widespread evaluation of germline and somatic profiling. Another GI malignancy with rich targeting opportunities is CCA, specifically ICC and increasing literature is emerging in this disease utilizing DDR strategies. Summing up, while these patient subsets are relatively uncommon, a broader group of patients with GI malignancies and underlying HR gene mutations beyond BRCA1/2 exist, and these patients need to be identified for potential syndrome identification, cascade family testing and treatment implications. It is now evidently clear that DNA-repair treatment approaches are established with the use of platinum therapies and PARPis, and we need to build on these early signals with novel combinations and expand the applicable patient population. A recent paper based on the National Cancer Database pointed out that even though clear superiority in terms of OS has been observed in patients enrolled in clinical trials, only 0.1% of patients are able to participate in clinical trials (136). Thus, a related key mission is expansion of the number of high priority clinical trials
To quote Robert Frost “And miles to go before I sleep, and miles to go before I sleep.”
Acknowledgments
Dr. Mary Elizabeth Paulk for her constant support and words of encouragement during the last few months.
Funding: David M. Rubenstein Center for Pancreatic Cancer Research, Reiss Family Foundation, Cancer Support Grant P30 CA008748.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Shaheenah Dawood) for the series “Targeting the DNA Damaging Pathway: PARPi and Beyond” published in Chinese Clinical Oncology. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cco-2019-ddp-05). The series “Targeting the DNA Damaging Pathway: PARPi and Beyond” was commissioned by the editorial office without any funding or sponsorship. Dr. EMO serves as an unpaid Associate Editor-in-Chief of Chinese Clinical Oncology from October 2019 to September 2021. EMO reports grants from Genentech-Roche, grants and personal fees from BMS-Celgene, grants from MabVax Therapeutics, grants from BioAtla, grants and personal fees from AstraZenica, grants from Silenseed, personal fees from CytomX Therapeutics, personal fees from Rafael Therapeutics, personal fees from BioLineRx, personal fees from Merck, grants and personal fees from Polaris, personal fees from Sobi, personal fees from Molecular Templates, grants and personal fees from Ipsen, during the conduct of the study. NP has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990;250:1684-9. [Crossref] [PubMed]
- Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995;378:789-92. [Crossref] [PubMed]
- White AW, Almassy R, Calvert AH, et al. Resistance-modifying agents. 9. Synthesis and biological properties of benzimidazole inhibitors of the DNA repair enzyme poly(ADP-ribose) polymerase. J Med Chem 2000;43:4084-97. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. [Crossref] [PubMed]
- Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017;317:2402-16. [Crossref] [PubMed]
- Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 1999;91:1310-6. [Crossref] [PubMed]
- Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst 2002;94:1358-65. [Crossref] [PubMed]
- Brose MS, Rebbeck TR, Calzone KA, et al. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst 2002;94:1365-72. [Crossref] [PubMed]
- Mersch J, Jackson MA, Park M, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2015;121:269-75. [Crossref] [PubMed]
- Noh JM, Choi DH, Baek H, et al. Associations between BRCA Mutations in High-Risk Breast Cancer Patients and Familial Cancers Other than Breast or Ovary. J Breast Cancer 2012;15:283-7. [Crossref] [PubMed]
- Yost S, Ruark E, Alexandrov LB, et al. Insights into BRCA Cancer Predisposition from Integrated Germline and Somatic Analyses in 7632 Cancers. JNCI Cancer Spectr 2019;3:pkz028. [Crossref] [PubMed]
- Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576-9. Erratum in: Nature. 2020 Jan;577(7789):E1. doi: 10.1038/s41586-019-1839-2. [Crossref] [PubMed]
- Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 2000;408:429-32. [Crossref] [PubMed]
- Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 2017;58:235-63. [Crossref] [PubMed]
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152-8. [Crossref] [PubMed]
- Betermier M, Bertrand P, Lopez BS. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet 2014;10:e1004086. [Crossref] [PubMed]
- Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell 2001;7:273-82. [Crossref] [PubMed]
- Zhao W, Steinfeld JB, Liang F, et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017;550:360-5. [Crossref] [PubMed]
- Reilly NM, Yard BD, Pittman DL. Homologous Recombination-Mediated DNA Repair and Implications for Clinical Treatment of Repair Defective Cancers. Methods Mol Biol 2019;1999:3-29. [Crossref] [PubMed]
- Byrum AK, Vindigni A, Mosammaparast N. Defining and Modulating 'BRCAness'. Trends Cell Biol 2019;29:740-51. [Crossref] [PubMed]
- Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 2014;20:764-75. [Crossref] [PubMed]
- Kondo T, Kanai M, Kou T, et al. Association between homologous recombination repair gene mutations and response to oxaliplatin in pancreatic cancer. Oncotarget 2018;9:19817-25. [Crossref] [PubMed]
- Thompson LH. Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res 2012;751:158-246. [Crossref] [PubMed]
- Dobzhansky T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics 1946;31:269-90. [PubMed]
- Kaelin WG Jr. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer 2005;5:689-98. [Crossref] [PubMed]
- Kedar PS, Stefanick DF, Horton JK, et al. Increased PARP-1 association with DNA in alkylation damaged, PARP-inhibited mouse fibroblasts. Mol Cancer Res 2012;10:360-8. [Crossref] [PubMed]
- Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature 1992;356:356-8. [Crossref] [PubMed]
- Ruscetti T, Lehnert BE, Halbrook J, et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem 1998;273:14461-7. [Crossref] [PubMed]
- Do K, Chen AP. Molecular pathways: targeting PARP in cancer treatment. Clin Cancer Res 2013;19:977-84. [Crossref] [PubMed]
- Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer 2004;4:814-9. [Crossref] [PubMed]
- Ledermann JA, Harter P, Gourley C, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol 2016;17:1579-89. [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. [Crossref] [PubMed]
- Pilié PG, Tang C, Mills GB, et al. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol 2019;16:81-104. [Crossref] [PubMed]
- Sulkowski PL, Corso CD, Robinson ND, et al. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med 2017;9:eaal2463.
- Cerrato A, Morra F, Celetti A. Use of poly ADP-ribose polymerase [PARP] inhibitors in cancer cells bearing DDR defects: the rationale for their inclusion in the clinic. J Exp Clin Cancer Res 2016;35:179. [Crossref] [PubMed]
- Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell 2011;19:664-78. [Crossref] [PubMed]
- Brenner JC, Feng FY, Han S, et al. PARP-1 inhibition as a targeted strategy to treat Ewing's sarcoma. Cancer Res 2012;72:1608-13. [PubMed]
- Joshi PM, Sutor SL, Huntoon CJ, et al. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 2014;289:9247-53. [PubMed]
- Postel-Vinay S, Bajrami I, Friboulet L, et al. A high-throughput screen identifies PARP1/2 inhibitors as a potential therapy for ERCC1-deficient non-small cell lung cancer. Oncogene 2013;32:5377-87. [PubMed]
- Yu H, Pak H, Hammond-Martel I, et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc Natl Acad Sci U S A 2014;111:285-90. [Crossref] [PubMed]
- Lu Y, Kwintkiewicz J, Liu Y, et al. Chemosensitivity of IDH1-Mutated Gliomas Due to an Impairment in PARP1-Mediated DNA Repair. Cancer Res 2017;77:1709-18. [Crossref] [PubMed]
- Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 1996;56:5360-4. [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Connor AA, Denroche RE, Jang GH, et al. Association of Distinct Mutational Signatures With Correlates of Increased Immune Activity in Pancreatic Ductal Adenocarcinoma. JAMA Oncol 2017;3:774-83. [Crossref] [PubMed]
- Lowery MA, Kelsen DP, Stadler ZK, et al. An emerging entity: pancreatic adenocarcinoma associated with a known BRCA mutation: clinical descriptors, treatment implications, and future directions. Oncologist 2011;16:1397-402. [Crossref] [PubMed]
- Lowery MA, Jordan EJ, Basturk O, et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin Cancer Res 2017;23:6094-100. [Crossref] [PubMed]
- Golan T, Kanji ZS, Epelbaum R, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111:1132-8. [Crossref] [PubMed]
- Kondo T, Kanai M, Kou T, et al. Impact of BRCAness on the efficacy of oxaliplatin-based chemotherapy in patients with unresectable pancreatic cancer. J Clin Oncol 2017;35:abstr 250.
- Golan T, Sella T, O'Reilly EM, et al. Overall survival and clinical characteristics of BRCA mutation carriers with stage I/II pancreatic cancer. Br J Cancer 2017;116:697-702. [Crossref] [PubMed]
- Fogelman DR, Wolff RA, Kopetz S, et al. Evidence for the efficacy of Iniparib, a PARP-1 inhibitor, in BRCA2-associated pancreatic cancer. Anticancer Res 2011;31:1417-20. [PubMed]
- LoRusso PM, Li J, Burger A, et al. Phase I Safety, Pharmacokinetic, and Pharmacodynamic Study of the Poly(ADP-ribose) Polymerase (PARP) Inhibitor Veliparib (ABT-888) in Combination with Irinotecan in Patients with Advanced Solid Tumors. Clin Cancer Res 2016;22:3227-37. [Crossref] [PubMed]
- Berlin J, Ramanathan RK, Strickler JH, et al. A phase 1 dose-escalation study of veliparib with bimonthly FOLFIRI in patients with advanced solid tumours. Br J Cancer 2018;118:938-46. [Crossref] [PubMed]
- Yarchoan M, Myzak MC, Johnson BA 3rd, et al. Olaparib in combination with irinotecan, cisplatin, and mitomycin C in patients with advanced pancreatic cancer. Oncotarget 2017;8:44073-81. [Crossref] [PubMed]
- Lowery MA, Kelsen DP, Capanu M, et al. Phase II trial of veliparib in patients with previously treated BRCA-mutated pancreas ductal adenocarcinoma. Eur J Cancer 2018;89:19-26. [Crossref] [PubMed]
- Shroff RT, Hendifar A, McWilliams RR, et al. Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. JCO Precis Oncol 2018;2018. [Crossref] [PubMed]
- O'Reilly EM, Lee JW, Lowery MA, et al. Phase 1 trial evaluating cisplatin, gemcitabine, and veliparib in 2 patient cohorts: Germline BRCA mutation carriers and wild-type BRCA pancreatic ductal adenocarcinoma. Cancer 2018;124:1374-82. [Crossref] [PubMed]
- O'Reilly EM, Lee JW, Zalupski M, et al. Randomized, Multicenter, Phase II Trial of Gemcitabine and Cisplatin With or Without Veliparib in Patients With Pancreas Adenocarcinoma and a Germline BRCA/PALB2 Mutation. J Clin Oncol 2020;38:1378-88. [Crossref] [PubMed]
- Golan T, Hammel P, Reni M, et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N Engl J Med 2019;381:317-27. [Crossref] [PubMed]
- Pant S, Maitra A, Yap TA. PARP inhibition - opportunities in pancreatic cancer. Nat Rev Clin Oncol 2019;16:595-6. [Crossref] [PubMed]
- Hammel P, Kindler HL, Reni M, et al. Health-related quality of life in patients with a germline BRCA mutation and metastatic pancreatic cancer receiving maintenance olaparib. Ann Oncol 2019;30:1959-68. [Crossref] [PubMed]
- Okur V, Chung WK. The impact of hereditary cancer gene panels on clinical care and lessons learned. Cold Spring Harb Mol Case Stud 2017;3:a002154. [Crossref] [PubMed]
- Syngal S, Furniss CS. Germline Genetic Testing for Pancreatic Ductal Adenocarcinoma at Time of Diagnosis. JAMA 2018;319:2383-5. [Crossref] [PubMed]
- Hu C, Hart SN, Polley EC, et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. JAMA 2018;319:2401-9. [Crossref] [PubMed]
- Pishvaian MJ, Bender RJ, Halverson D, et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin Cancer Res 2018;24:5018-27. [Crossref] [PubMed]
- Pishvaian MJ, Blais EM, Brody JR, et al. Outcomes in Patients with Pancreatic Adenocarcinoma With Genetic Mutations in DNA Damage Response Pathways: Results From the Know Your Tumor Program. JCO Precis Oncol 2019. [Crossref]
- Park W, Wong W, Yu KH, et al. Homologous recombination deficiency (HRD): A biomarker for first-line (1L) platinum in advanced pancreatic ductal adenocarcinoma (PDAC). J Clin Oncol 2019;37:abstr 4132.
- Park W, Chen J, Chou JF, et al. Genomic Methods Identify Homologous Recombination Deficiency in Pancreas Adenocarcinoma and Optimize Treatment Selection. Clin Cancer Res 2020;26:3239-47. [Crossref] [PubMed]
- Chiorean EG, Guthrie KA, Philip PA, et al. Randomized phase II study of second-line modified FOLFIRI with PARP inhibitor ABT-888 (Veliparib) (NSC-737664) versus FOLFIRI in metastatic pancreatic cancer (mPC): SWOG S1513. J Clin Oncol 2019;37:abstr 4014.
- Pishvaian MJ, Wang H, Parenti S, et al. Final report of a phase I/II study of veliparib (Vel) in combination with 5-FU and oxaliplatin (FOLFOX) in patients (pts) with metastatic pancreatic cancer (mPDAC). J Clin Oncol 2019;37:abstr 4015.
- Hu N, Wang C, Han XY, et al. Evaluation of BRCA2 in the genetic susceptibility of familial esophageal cancer. Oncogene 2004;23:852-8. [Crossref] [PubMed]
- Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014;5:2839-52. [Crossref] [PubMed]
- Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47:1003-10. [Crossref] [PubMed]
- Churi CR, Shroff R, Wang Y, et al. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PLoS One 2014;9:e115383. [Crossref] [PubMed]
- Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470-3. [Crossref] [PubMed]
- Al-Shamsi HO, Anand D, Shroff RT, et al. BRCA-associated protein 1 mutant cholangiocarcinoma: an aggressive disease subtype. Journal of Gastrointestinal Oncology 2016;7:556-61. [Crossref] [PubMed]
- Hirosawa T, Ishida M, Ishii K, et al. Loss of BAP1 expression is associated with genetic mutation and can predict outcomes in gallbladder cancer. PLoS One 2018;13:e0206643. [Crossref] [PubMed]
- Lin J, Shi J, Guo H, et al. Alterations in DNA Damage Repair Genes in Primary Liver Cancer. Clin Cancer Res 2019;25:4701-11. [Crossref] [PubMed]
- Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive Molecular Profiling of Intrahepatic and Extrahepatic Cholangiocarcinomas: Potential Targets for Intervention. Clin Cancer Res 2018;24:4154-61. [Crossref] [PubMed]
- Parasramka M, Yan IK, Wang X, et al. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer 2017;16:22. [Crossref] [PubMed]
- Golan T, Raitses-Gurevich M, Kelley RK, et al. Overall Survival and Clinical Characteristics of BRCA-Associated Cholangiocarcinoma: A Multicenter Retrospective Study. Oncologist 2017;22:804-10. [Crossref] [PubMed]
- Mao Y, Huang X, Shuang Z, et al. PARP inhibitor olaparib sensitizes cholangiocarcinoma cells to radiation. Cancer Med 2018;7:1285-96. [Crossref] [PubMed]
- Akbari MR, Malekzadeh R, Nasrollahzadeh D, et al. Germline BRCA2 mutations and the risk of esophageal squamous cell carcinoma. Oncogene 2008;27:1290-6. [Crossref] [PubMed]
- Moran A, O'Hara C, Khan S, et al. Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam Cancer 2012;11:235-42. [PubMed]
- Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 2016;48:1131-41. [Crossref] [PubMed]
- Deng J, Weng X, Ye J, et al. Identification of the Germline Mutation Profile in Esophageal Squamous Cell Carcinoma by Whole Exome Sequencing. Front Genet 2019;10:47. [Crossref] [PubMed]
- Miyamoto K, Minegaki T, Tanahashi M, et al. Synergistic Effects of Olaparib and DNA-damaging Agents in Oesophageal Squamous Cell Carcinoma Cell Lines. Anticancer Res 2019;39:1813-20. [Crossref] [PubMed]
- Evans HS, Lewis CM, Robinson D, et al. Incidence of multiple primary cancers in a cohort of women diagnosed with breast cancer in southeast England. Br J Cancer 2001;84:435-40. [Crossref] [PubMed]
- Risch HA, McLaughlin JR, Cole DE, et al. Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 2001;68:700-10. [Crossref] [PubMed]
- Kim JW, Cho HJ, Kim M, et al. Differing effects of adjuvant chemotherapy according to BRCA1 nuclear expression in gastric cancer. Cancer Chemother Pharmacol 2013;71:1435-43. [Crossref] [PubMed]
- Chen W, Wang J, Li X, et al. Prognostic significance of BRCA1 expression in gastric cancer. Med Oncol 2013;30:423. [Crossref] [PubMed]
- Yan S, He F, Luo R, et al. Decreased expression of BRCA1-associated protein 1 predicts unfavorable survival in gastric adenocarcinoma. Tumour Biol 2016;37:6125-33. [Crossref] [PubMed]
- Kim HS, Hwang IG, Min HY, et al. Clinical significance of BRCA1 and BRCA2 mRNA and protein expression in patients with sporadic gastric cancer. Oncol Lett 2019;17:4383-92. [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Mihailidou C, Karamouzis MV, Schizas D, et al. Co-targeting c-Met and DNA double-strand breaks (DSBs): Therapeutic strategies in BRCA-mutated gastric carcinomas. Biochimie 2017;142:135-43. [Crossref] [PubMed]
- Zhang ZZ, Liu YJ, Yin XL, et al. Loss of BRCA1 expression leads to worse survival in patients with gastric carcinoma. World J Gastroenterol 2013;19:1968-74. [Crossref] [PubMed]
- Wang GH, Zhao CM, Huang Y, et al. BRCA1 and BRCA2 expression patterns and prognostic significance in digestive system cancers. Hum Pathol 2018;71:135-44. [Crossref] [PubMed]
- Kim G, Kim J, Han SY, et al. The effects of BRCA1 expression on the chemosensitivity of gastric cancer cells to platinum agents. Oncol Lett 2019;17:5023-9. [PubMed]
- Kubota E, Williamson CT, Ye R, et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 2014;13:2129-37. [Crossref] [PubMed]
- Bang YJ, Im SA, Lee KW, et al. Randomized, Double-Blind Phase II Trial With Prospective Classification by ATM Protein Level to Evaluate the Efficacy and Tolerability of Olaparib Plus Paclitaxel in Patients With Recurrent or Metastatic Gastric Cancer. J Clin Oncol 2015;33:3858-65. [Crossref] [PubMed]
- Bang YJ, Xu RH, Chin K, et al. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:1637-51. [Crossref] [PubMed]
- Yang L, Yang G, Ding Y, et al. Combined treatment with PI3K inhibitor BKM120 and PARP inhibitor olaparib is effective in inhibiting the gastric cancer cells with ARID1A deficiency. Oncol Rep 2018;40:479-87. [PubMed]
- Yurgelun MB, Kulke MH, Fuchs CS, et al. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. J Clin Oncol 2017;35:1086-95. [Crossref] [PubMed]
- Phelan CM, Iqbal J, Lynch HT, et al. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: results from a follow-up study. Br J Cancer 2014;110:530-4. [Crossref] [PubMed]
- Kirchhoff T, Satagopan JM, Kauff ND, et al. Frequency of BRCA1 and BRCA2 mutations in unselected Ashkenazi Jewish patients with colorectal cancer. J Natl Cancer Inst 2004;96:68-70. [Crossref] [PubMed]
- Niell BL, Rennert G, Bonner JD, et al. BRCA1 and BRCA2 founder mutations and the risk of colorectal cancer. J Natl Cancer Inst 2004;96:15-21. [Crossref] [PubMed]
- Oh M, McBride A, Yun S, et al. BRCA1 and BRCA2 Gene Mutations and Colorectal Cancer Risk: Systematic Review and Meta-analysis. J Natl Cancer Inst 2018;110:1178-89. [Crossref] [PubMed]
- Parikh AR, He Y, Hong TS, et al. Analysis of DNA Damage Response Gene Alterations and Tumor Mutational Burden Across 17,486 Tubular Gastrointestinal Carcinomas: Implications for Therapy. Oncologist 2019;24:1340-7. [Crossref] [PubMed]
- Davidson D, Wang Y, Aloyz R, et al. The PARP inhibitor ABT-888 synergizes irinotecan treatment of colon cancer cell lines. Invest New Drugs 2013;31:461-8. [Crossref] [PubMed]
- Shelton JW, Waxweiler TV, Landry J, et al. In vitro and in vivo enhancement of chemoradiation using the oral PARP inhibitor ABT-888 in colorectal cancer cells. Int J Radiat Oncol Biol Phys 2013;86:469-76. [Crossref] [PubMed]
- Wang C, Jette N, Moussienko D, et al. ATM-Deficient Colorectal Cancer Cells Are Sensitive to the PARP Inhibitor Olaparib. Transl Oncol 2017;10:190-6. [Crossref] [PubMed]
- Leichman L, Groshen S, O'Neil BH, et al. Phase II Study of Olaparib (AZD-2281) After Standard Systemic Therapies for Disseminated Colorectal Cancer. Oncologist 2016;21:172-7. [Crossref] [PubMed]
- Pishvaian MJ, Slack RS, Jiang W, et al. A phase 2 study of the PARP inhibitor veliparib plus temozolomide in patients with heavily pretreated metastatic colorectal cancer. Cancer 2018;124:2337-46. [Crossref] [PubMed]
- Gorbunova V, Beck JT, Hofheinz RD, et al. A phase 2 randomised study of veliparib plus FOLFIRI+/-bevacizumab versus placebo plus FOLFIRI+/-bevacizumab in metastatic colorectal cancer. Br J Cancer 2019;120:183-9. [Crossref] [PubMed]
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110-20. [Crossref] [PubMed]
- Curtin NJ, Drew Y, Sharma-Saha S. Why BRCA mutations are not tumour-agnostic biomarkers for PARP inhibitor therapy. Nat Rev Clin Oncol 2019;16:725-6. [Crossref] [PubMed]
- Criscuolo D, Morra F, Giannella R, et al. Identification of Novel Biomarkers of Homologous Recombination Defect in DNA Repair to Predict Sensitivity of Prostate Cancer Cells to PARP-Inhibitors. Int J Mol Sci 2019;20:3100. [Crossref] [PubMed]
- Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008;451:1116-20. [Crossref] [PubMed]
- Chan N, Pires IM, Bencokova Z, et al. Contextual synthetic lethality of cancer cell kill based on the tumor microenvironment. Cancer Res 2010;70:8045-54. [Crossref] [PubMed]
- Zhan L, Qin Q, Lu J, et al. Novel poly (ADP-ribose) polymerase inhibitor, AZD2281, enhances radiosensitivity of both normoxic and hypoxic esophageal squamous cancer cells. Dis Esophagus 2016;29:215-23. [Crossref] [PubMed]
- Liu JF, Barry WT, Birrer M, et al. Combination cediranib and olaparib versus olaparib alone for women with recurrent platinum-sensitive ovarian cancer: a randomised phase 2 study. Lancet Oncol 2014;15:1207-14. [Crossref] [PubMed]
- Parkes EE, Walker SM, Taggart LE, et al. Activation of STING-Dependent Innate Immune Signaling By S-Phase-Specific DNA Damage in Breast Cancer. J Natl Cancer Inst 2016;109:djw199. [Crossref] [PubMed]
- Teo MY, Seier K, Ostrovnaya I, et al. Alterations in DNA Damage Response and Repair Genes as Potential Marker of Clinical Benefit From PD-1/PD-L1 Blockade in Advanced Urothelial Cancers. J Clin Oncol 2018;36:1685-94. [Crossref] [PubMed]
- Brown JS, Sundar R, Lopez J. Combining DNA damaging therapeutics with immunotherapy: more haste, less speed. Br J Cancer 2018;118:312-24. [Crossref] [PubMed]
- Lee JM, Cimino-Mathews A, Peer CJ, et al. Safety and Clinical Activity of the Programmed Death-Ligand 1 Inhibitor Durvalumab in Combination With Poly (ADP-Ribose) Polymerase Inhibitor Olaparib or Vascular Endothelial Growth Factor Receptor 1-3 Inhibitor Cediranib in Women's Cancers: A Dose-Escalation, Phase I Study. J Clin Oncol 2017;35:2193-202. [Crossref] [PubMed]
- Lok BH, Carley AC, Tchang B, et al. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 2013;32:3552-8. [Crossref] [PubMed]
- Huang F, Goyal N, Sullivan K, et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic Acids Res 2016;44:4189-99. [Crossref] [PubMed]
- Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood 2013;122:1293-304. [Crossref] [PubMed]
- Hengel SR, Malacaria E, Folly da Silva Constantino L, et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. Elife 2016;5:e14740. [Crossref] [PubMed]
- Jin J, Fang H, Yang F, et al. Combined Inhibition of ATR and WEE1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Neoplasia 2018;20:478-88. [Crossref] [PubMed]
- Karnak D, Engelke CG, Parsels LA, et al. Combined inhibition of Wee1 and PARP1/2 for radiosensitization in pancreatic cancer. Clin Cancer Res 2014;20:5085-96. [Crossref] [PubMed]
- Mateos-Gomez PA, Gong F, Nair N, et al. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015;518:254-7. [Crossref] [PubMed]
- Malapelle U, Morra F, Ilardi G, et al. USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs. Lung Cancer 2017;107:41-9. [Crossref] [PubMed]
- Yap TA, Plummer R, Azad NS, et al. The DNA Damaging Revolution: PARP Inhibitors and Beyond. Am Soc Clin Oncol Educ Book 2019;39:185-95. [Crossref] [PubMed]
- Zaorsky NG, Zhang Y, Walter V, et al. Clinical Trial Accrual at Initial Course of Therapy for Cancer and Its Impact on Survival. J Natl Compr Canc Netw 2019;17:1309-16. [Crossref] [PubMed]


