BRCA sequencing of tumors: understanding its implications in the oncology community
Introduction
The breast cancer susceptibility genes BRCA1/2 are tumor suppressor genes, that when mutated, increases an individual’s risk of developing not just breast cancer, but also ovarian, pancreatic, and prostate cancer (1-4). This occurs as the BRCA1/2 genes encode for proteins that play crucial roles in the maintenance of genomic integrity by ensuring accurate and precise repair of damaged DNA, as well as controlling cell cycle checkpoints (5,6). Women who harbor pathogenic BRCA1/2 mutations are at a 45–65% risk of developing breast cancer and a 11–39% risk of ovarian cancer by the age of 70 (7), compared to the corresponding general population cumulative risk of 5.03% and 0.72% respectively (8). The past 25 years have seen an increase in the number of genetic tests being offered, owing to advances in testing technology and increase in access to testing (9). With lowered test costs and high-throughput sequencing technologies, many more laboratories have been able to offer tests that cover more genes, beyond the conventional BRCA1/2 genes, to diagnose hereditary breast cancer syndrome. Indeed, the rates of testing for hereditary breast cancer including BRCA1/2 have been increasing over the years (10,11). This has led to a need to address the implications of acquiring genetic information; be it intentionally sought or incidentally discovered. In this review article, we seek to provide a brief history of DNA sequencing, highlight the differences between sequencing DNA from blood and tumor samples, with particular emphasis on the BRCA1/2 genes, as well as understand the clinical implications of tumor BRCA1/2 testing.
The process of DNA sequencing and its utility
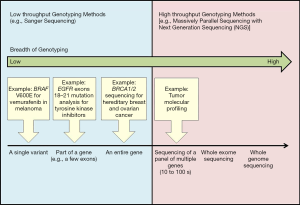
DNA sequencing was first described in the late 1970s by Fred Sanger and his colleagues (12) and is better known as the Sanger sequencing method. Sanger sequencing involves mixing non-extendable, fluorescently labeled dideoxy nucleotides together with standard nucleotides that may be randomly incorporated by DNA polymerase to generate fragments of varying lengths of nucleotides which are copies of the original template of DNA. These fragments are then separated by high-resolution capillary electrophoresis, and the color tag linked to the last incorporated dideoxy nucleotide on each fragment is then used to interpret the original sequence of DNA (13). It was the Sanger sequencing method which was employed to elucidate the first complete sequence of the human genome (known as the Human Genome Project) (14). However, the traditional Sanger sequencing method is limited by its throughput as well as high cost. In fact, it was estimated that the first human genome sequencing cost an estimated 0.5–1 billion US dollars (15). Since then, much effort has been poured into initiatives to improve DNA sequencing, in a bid to increase throughput whilst reducing cost. The National Human Genome Research Institute created a 70-million-dollar DNA sequencing technology initiative with the aim of achieving a 1,000-dollar human genome within 10 years (16). One such technology that was created is known as next-generation sequencing (NGS). While the concept behind Sanger sequencing and NGS is similar, the critical difference lies in the sequencing volume. The traditional Sanger sequencing method sequences a single DNA fragment at any one time, while NGS, also known as massive parallel sequencing, sequences millions of fragments simultaneously per run. This capability has expanded the breadth of genotyping available in the clinic. We can now utilize different technologies to sequence genetic information of varying lengths (17). Low throughput methods such as Sanger sequencing are suitably used to sequence single variants such as the BRAF V600E mutation, part of a gene such as epidermal growth factor receptor (EGFR) mutations in exons 18–21 (18), or an entire gene for example full BRCA1/2 sequencing. At the other end of the spectrum, high-throughput genotyping such as NGS method has the capability of sequencing tens to hundreds of genes simultaneously, the whole exome, or even the whole genome (Figure 1).
After a genomic sequence is determined, the reads are aligned to a published reference genome and compared. Any site with a differing DNA from the reference is considered a sequence variant (19). Variants are classified as “pathogenic”, “likely pathogenic”, “of uncertain significance”, “likely benign” or “benign” based on the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology guidelines (20). By increasing the length of genome that is sequenced, the chance of detecting informative and/or actionable mutations is increased; however, the disadvantages of testing a large panel of genes include higher rates of variants of uncertain significance (VUS) (21) which are uninformative and not actionable, as well as a higher likelihood of incidental and/or unexpected findings in less familiar genes that may have limited or no management guidelines.
Germline vs. somatic testing (Table 1)

Full table
Germline mutations are heritable and present in every cell of a person’s body. These mutations are inherited from a person’s parent(s), and can be passed on to his or her offspring. Typical starting materials used to diagnose a germline mutation are blood (peripheral blood mononuclear cells), buccal swab, or saliva. Heritable germline mutations exist from birth to death and do not change with time, thus testing only needs to be done once. Heritable germline information can potentially lead to ethical, social, and familial implications, and hence pre-test genetic counseling is required. In oncology, the traditional indications for germline testing are to diagnose hereditary cancer syndrome, such as BRCA1/2 hereditary breast-ovarian cancer syndrome, to identify individuals who will benefit from early screening and prevention strategies (22,23). More recently, germline BRCA1/2 testing may lead to therapeutic indications, as germline BRCA1/2 mutation carriers with metastatic HER2 negative breast, advanced epithelial ovarian, or metastatic pancreatic cancer may benefit from treatment with poly-ADP-ribose polymerase (PARP) inhibitors (3,24-26). In contrast to germline mutations, somatic mutations are acquired genetic changes that occur in a diseased organ, for example, in tumor cells in a patient with cancer. Somatic testing in cancer is typically done on tumor biopsy or surgical specimens, or any other biological materials that contain malignant cells, e.g., malignant pleural effusion or ascitic fluid. As somatic mutations are not heritable, pre-test genetic counselling is generally not required. The traditional indication to test for somatic mutations in oncology is for therapeutics, e.g., identification of EGFR mutations in non-small cell lung cancer (NSCLC) to select patients for treatment with EGFR tyrosine kinase inhibitors (TKIs) (18,27,28). Somatic mutations may evolve with time and treatment exposure, thus repeated testing may be required, for example, testing for the emergence of EGFR T790M mutation in EGFR-mutant NSCLC patients who have developed resistance to first- or second-generation EGFR-TKIs (29). While genetic testing of blood is usually associated with germline information and testing of tumor with somatic information, this relationship does not always hold true. For example, although tumor is not commonly used as a source of germline DNA, it originates from normal tissue and thus contains germline DNA. Therefore, tumor genetic testing can potentially yield incidental germline findings. Similarly, if blood is processed to yield circulating tumor cells or cell-free DNA, then testing such materials will provide somatic rather than germline information.
Testing for mutations in the BRCA1/2 gene in the tumor
Germline testing for the BRCA1/2 gene is usually done on a blood sample. However, there are instances whereby BRCA1/2 gene sequencing is performed on a tumor sample. One scenario that is occurring increasingly commonly is when the tumor of a patient with refractory cancer is profiled using NGS in search of an actionable mutation to guide treatment. A panel of tens to several hundred genes is typically tested and often includes the BRCA1/2 genes. Pathogenic BRCA1/2 gene mutation may be identified in the tumor, and although some of these mutations are somatic mutations, others may represent incidental germline findings. Another example is when there is a deliberate search for a tumor BRCA1/2 mutation to guide treatment, for example in the setting of epithelial ovarian cancer, where there is data to support the use of PARP inhibitors in patients whose tumors harbor BRCA1/2 mutation (3,24), regardless of whether they are germline or somatic in nature (30). Lastly and rarely, tumor may be used as a surrogate for germline testing in the context of a high-risk family with no living affected index patient for direct germline testing.
Are BRCA1/2 gene mutations identified on tumor sequencing somatic or germline in origin?
One challenge when a pathogenic BRCA1/2 mutation is identified in tumor is to ascertain its origin, i.e., germline versus somatic. The most conclusive way to determine if a mutation identified in a tumor is germline or somatic in nature is to test an accompanying germline sample (e.g., blood or buccal swab). However, in practice, most laboratories offering tumor NGS testing do not routinely request for a germline sample from the patient, for several reasons. These include ethical concerns, as testing germline samples yields direct information on heritable mutations and will require prior genetic counselling, and practical considerations, such as the need for more bioinformatics analysis with a germline sample, thus increasing testing cost and possibly turnaround time (31). Most tumor NGS reports are silent on whether an identified pathogenic mutation is germline or somatic in origin. However, certain features may help to ascertain if a tumor variant could actually be germline in nature, including a concordant clinical and family history and mutant allele frequency (MAF). When a pathogenic BRCA1/2 mutation is identified in tumor, the clinician should review the patient’s clinical presentation to determine if it is consistent with a heritable mutation. Information such as the cancer type, family history and young age at diagnosis are important clues that could point towards the fact that the identified mutation may be germline in nature. Another clue is the MAF of the pathogenic variant of interest. Since most hereditary cancers are inherited in an autosomal dominant fashion (50% wild type and 50% mutant), the MAF of pathogenic germline variants is usually close to 50%, whereas MAF of pathogenic somatic variants tends to be much more variable. The gene in which the tumor pathogenic mutation is identified may provide further insights on its origin. For example, Meric-Bernstam et al. showed that the majority (77.8%) of tumor pathogenic BRCA1/2 variant was germline in nature, compared to only 2.88% of tumor pathogenic TP53 variants (32). Other clues that could suggest a tumor pathogenic mutation to be germline in nature include detection of the same mutation in different primary tumor specimens from the same patient, and the mutation having been previously reported as a heritable founder mutation (31).
What is the likelihood of finding an incidental germline pathogenic mutation on tumor NGS testing?
In a study from MD Anderson, 1,000 cancer patients underwent tumor NGS testing with a panel of 202 genes; all patients provided a corresponding germline sample (blood or buccal swab), allowing investigators to ascertain if an identified tumor pathogenic mutation is germline or somatic in nature. A focused analysis on 19 cancer predisposing genes found that ~5% of pathogenic mutations identified in the tumor in these 19 genes were actually germline in nature (31). Schrader et al. further reported that the likelihood of picking up incidental germline pathogenic mutations in tumor increases with the number of genes tested, from 6.4% in a 26-gene panel to 12.6% using a 93-gene panel and further to 15.7% when a 187-gene panel is tested (33). These mutations may be linked to increased risk of preventable diseases for which clear management guidelines are available (32), while others may cause non-preventable diseases or have less certain clinical implications. These findings have potential medico-legal implications, and there has been significant debate on whether all or some of these incidental germline findings should be disclosed to the patient. Importantly, disclosing and managing all such results demand significant medical expertise and health resources, which are not available at most institutions. Furthermore, disclosure of a finding that can lead to a non-preventable disease can also be a source of distress and may not be welcome by the patient. Physicians ordering these tests should thus be mindful of these issues.
How should we handle incidental BRCA1/2 pathogenic germline variants identified in tumor?
The ACMG recommends a list of 59 genes including 25 cancer predisposing genes in which results should be returned to patients if incidental germline mutations are identified; regardless of the original indication for the clinical sequencing (34). This includes the BRCA1/2 genes (34). Several studies have reported the likelihood of detecting an incidental germline pathogenic BRCA1/2 mutation on tumor NGS testing to range from 2.1% to 3.3% (32,33,35). Interestingly, the clinical diagnosis was not suspected in 24% to 46% of the cases. Clinicians should be aware of this when ordering tumor NGS testing for their patients; it will be appropriate to provide brief pre-test counseling to inform patients of the possibility of detecting incidental heritable mutations. If a tumor BRCA1/2 pathogenic mutation is identified, full genetic counseling should be provided with a view to confirm the mutation with clinical testing using a direct germline sample such as blood or buccal swab. Full genetic counselling typically comprises pre-and post-test processes. Pre-test genetic counselling includes taking a thorough family history for risk assessment followed by counselling on the characteristics of the suspected hereditary cancer syndrome, including mode of inheritance, lifetime cancer risks, screening and preventive options for proven mutation carriers, as well as highlighting potential ethical, social, and legal implications of genetic information. During post-test counselling, the implications of the test results are explained and follow-up plans including screening and prevention as well as cascade testing of family members of proven mutation carriers are formulated (22).
Is tumor BRCA1/2 gene testing reliable enough to diagnose germline mutations?
Since tumor NGS testing may uncover incidental germline BRCA1/2 mutations, an important clinical question arises: how reliable is tumor BRCA1/2 testing in diagnosing or excluding germline mutations? We studied 60 patients who had undergone clinical BRCA1/2 germline testing using blood samples, including 22 patients who were diagnosed with pathogenic germline BRCA1/2 mutations and 38 patients without (36). Paraffin-embedded tumors from these patients were retrieved for tumor BRCA1/2 testing via NGS. The laboratory was blinded to the germline test results and was asked to commit if an identified tumor pathogenic BRCA1/2 is germline in origin, using MAF and other variant classification algorithms. In the 38 patients with no germline pathogenic BRCA1/2 mutation, tumor tests were 100% concordant with no false positive results. However, in patients who carry germline pathogenic BRCA1/2 mutation (n=22), only 70% of these germline mutations were conclusively diagnosed on tumor testing, while 30% of germline mutations were missed. Among the false negative cases, 40% was due to technical error, i.e., the mutation was not detected in the tumor, while 60% was due to interpretative error, i.e., mutation was detected in the tumor but the classification algorithm used by the laboratory erroneously classified the variant as non-pathogenic or somatic. These results highlight that a tumor NGS test that is negative for BRCA1/2 mutation does not conclusively exclude germline pathogenic BRCA1/2 mutation, thus germline testing should still be considered if the patient fulfills clinical criteria for germline BRCA1/2 testing. On the other hand, tumor BRCA1/2 testing can detect some pathogenic germline mutations and false positive rates appear low, with high concordance rate of more than 90% between tumor BRCA1/2 mutations assessed to be likely germline in nature versus blood testing (37-39). Thus, in the context of a high risk family with no living affected who can be tested as the index patient, archival tumor specimen from a deceased cancer-affected family member can potentially be used as a surrogate for germline; the detection of a pathogenic germline mutation in this context could potentially facilitate cascade testing in family members, although this is best done in the context of a specialized cancer genetics clinic.
Putting it all together (Figure 2)
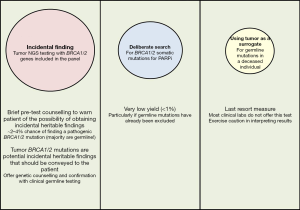
When sequencing a tumor using NGS testing, the probability of finding an incidental pathogenic BRCA1/2 mutation is ~2–4%, of which ~80% is germline in origin. Consequently, clinicians should consider pre-test counselling to warn patients of such a possibility (32). Subsequently, when a pathogenic BRCA1/2 gene mutation is indeed reported in the tumor, the patient should be counseled that this may represent an incidental germline finding. It is recommended that these patients be referred for formal genetic counselling followed by confirmatory germline testing with a blood sample or buccal swab. On the other hand, due to a relatively high false negative rate, failure to identify pathogenic BRCA1/2 mutation on tumor NGS testing does not definitively exclude germline BRCA1/2 mutations. Therefore, if an individual fulfills conventional genetic testing criteria, germline testing will still be indicated even if the tumor testing returns negative. Finally, using tumor as a surrogate to test for germline mutations in a deceased individual should only be used as a last resort, best done in the context of a cancer genetics clinic, and results interpreted with caution.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Shaheenah Dawood) for the series “Targeting the DNA Damaging Pathway: PARPi and Beyond” published in Chinese Clinical Oncology. The article was sent for external peer review organized by the Guest Editor and the editorial office.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/cco-19-198). The series “Targeting the DNA Damaging Pathway: PARPi and Beyond” was commissioned by the editorial office without any funding or sponsorship. Dr. SCL reports grants and other from Pfizer, other from Novartis, other from Astra Zeneca, grants and other from Taiho, grants and other from Eisai, other from Roche, grants and other from ACT Genomics, grants and other from ASLAN Pharmaceuticals, other from MSD, other from Amgen, other from Eli Lilly, outside the submitted work. RSJW has no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Armstrong N, Ryder S, Forbes C, et al. A systematic review of the international prevalence of BRCA mutation in breast cancer. Clin Epidemiol 2019;11:543-61. [Crossref] [PubMed]
- Thompson D, Easton DF. Cancer Incidence in BRCA1 Mutation Carriers. J Natl Cancer Inst 2002;94:1358-65. [Crossref] [PubMed]
- Paul A, Paul S. The breast cancer susceptibility genes (BRCA) in breast and ovarian cancers. Front Biosci (Landmark Ed) 2014;19:605-18. [Crossref] [PubMed]
- Cavanagh H, Rogers KMA. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hered Cancer Clin Pract 2015;13:16. [Crossref] [PubMed]
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006;25:5864-74. [Crossref] [PubMed]
- Orr KS, Savage KI. The BRCA1 and BRCA2 Breast and Ovarian Cancer Susceptibility Genes — Implications for DNA Damage Response, DNA Repair and Cancer Therapy. In: Chen CC. editor. Advances in DNA Repair. IntechOpen, November 18th 2015. doi: 10.5772/59996. [Crossref]
- Antoniou A, Pharoah PD, Narod S, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet 2003;72:1117-30. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Toland AE, Forman A, Couch FJ, et al. Clinical testing of BRCA1 and BRCA2: a worldwide snapshot of technological practices. NPJ Genom Med 2018;3:7. [Crossref] [PubMed]
- Rosenberg SM, Ruddy KJ, Tamimi RM, et al. BRCA1 and BRCA2 mutation testing in young women with breast cancer. JAMA Oncol 2016;2:730-6. [Crossref] [PubMed]
- Chen Z, Kolor K, Grosse SD, et al. Trends in utilization and costs of BRCA testing among women aged 18–64 years in the United States, 2003–2014. Genet Med 2018;20:428-34. [Crossref] [PubMed]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 1977;74:5463-7. [Crossref] [PubMed]
- Sanger F, Coulson AR. A rapid method for determining sequences of DNA by primed synthesis with DNA polymerase. J Mol Biol 1975;94:441-8. [Crossref] [PubMed]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature 2001;409:860-921. Erratum in: Nature 2001 Aug 2;412(6846):565; Nature 2001 Jun 7;411(6838):720. Szustakowki, J [corrected to Szustakowski, J]. [Crossref] [PubMed]
- Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell 2015;58:586-97. [Crossref] [PubMed]
- Schloss JA. How to get genomes at one ten-thousandth the cost. Nat Biotechnol 2008;26:1113-5. [Crossref] [PubMed]
- Caspar SM, Dubacher N, Kopps AM, et al. Clinical sequencing: From raw data to diagnosis with lifetime value. Clin Genet 2018;93:508-19. [Crossref] [PubMed]
- Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 2007;25:587-95. [Crossref] [PubMed]
- Bombard Y, Robson M, Offit K. Revealing the incidentalome when targeting the tumor genome. JAMA 2013;310:795-6. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Cheon JY, Mozersky J, Cook-Deegan R. Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med 2014;6:121. [Crossref] [PubMed]
- Moyer VA. U.S. Preventive Services Task Force. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:271-81. [PubMed]
- Balmaña J, Díez O, Rubio IT, et al. BRCA in breast cancer: ESMO Clinical Practice Guidelines. Ann Oncol 2011;22 Suppl 6:vi31-4. [Crossref] [PubMed]
- Przybycinski J, Nalewajska M, Marchelek-Mysliwiec M, et al. Poly-ADP-ribose polymerases (PARPs) as a therapeutic target in the treatment of selected cancers. Expert Opin Ther Targets 2019;23:773-85. [Crossref] [PubMed]
- Lee JM, Ledermann JA, Kohn EC. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann Oncol 2014;25:32-40. [Crossref] [PubMed]
- Tung NM, Garber JE. BRCA1/2 testing: therapeutic implications for breast cancer management. Br J Cancer 2018;119:141-52. [Crossref] [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Morgensztern D, Politi K, Herbst RS. EGFR Mutations in Non-Small-Cell Lung Cancer: Find, Divide, and Conquer. JAMA Oncol 2015;1:146-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Moschetta M, George A, Kaye SB, et al. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol 2016;27:1449-55. [Crossref] [PubMed]
- Raymond VM, Gray SW, Roychowdhury S, et al. Germline Findings in Tumor-Only Sequencing: Points to Consider for Clinicians and Laboratories. J Natl Cancer Inst 2015;108:djv351. [Crossref] [PubMed]
- Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol 2016;27:795-800. [Crossref] [PubMed]
- Schrader KA, Cheng DT, Joseph V, et al. Germline Variants in Targeted Tumor Sequencing Using Matched Normal DNA. JAMA Oncol 2016;2:104-11. Erratum in: JAMA Oncol. 2016 Feb;2(2):279. doi: 10.1001/jamaoncol.2015.6541. [Crossref] [PubMed]
- Kalia SS, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017;19:249-55. Erratum in: Genet Med. 2017 Apr;19(4):484. doi: 10.1038/gim.2017.17. [Crossref] [PubMed]
- Seifert BA, O'Daniel JM, Amin K, et al. Germline Analysis from Tumor-Germline Sequencing Dyads to Identify Clinically Actionable Secondary Findings. Clin Cancer Res 2016;22:4087-94. [Crossref] [PubMed]
- Ong PY, Poon SL, Tan KT, et al. Using next-generation sequencing (NGS) platform to diagnose pathogenic germline BRCA1/2 mutations from archival tumor specimens. Gynecol Oncol 2019;155:275-9. [Crossref] [PubMed]
- Hodgson DR, Dearden SP, Brown JS, et al. Analysis of tumor samples from SOLO2: Concordance of BRCA mutation (BRCAm) detection in tumor vs. blood and frequency of BRCA-specific loss of heterozygosity (LOH) and loss of function somatic mutations. J Clin Oncol 2018;36:abstr 12017.
- Gornjec A, Novakovic S, Stegel V, et al. Cytology material is equivalent to tumor tissue in determining mutations of BRCA 1/2 genes in patients with tubo-ovarian high grade serous carcinoma. BMC Cancer 2019;19:296. [Crossref] [PubMed]
- Dougherty BA, Lai Z, Hodgson DR, et al. Biological and clinical evidence for somatic mutations in BRCA1 and BRCA2 as predictive markers for olaparib response in high-grade serous ovarian cancers in the maintenance setting. Oncotarget 2017;8:43653-61. [Crossref] [PubMed]