Integrative oncology drug discovery accompanied by preclinical translational research as prerequisite for clinical development
Recent changes in oncology drug discovery and drug development
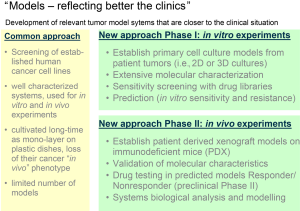
Looking back on more than 60 years of drug development for cancer therapy, almost in parallel with the new millennium, processes have changed substantially. This has been driven by increasing costs for the clinical development in contrast to often disappointing improvements for the patients. For more than 50 years, new cancer drugs were characterized in a handful of lowly predictive preclinical tumor models—and all further development work and risks were left to clinicians and patients. Growing insight into the fundamental genetic basics of the disease through analysis of gene expression and mutations and the development of fascinating new technologies in genetic engineering and bioinformatics—key word systems biology—have provided the technical basis for this paradigm shift.
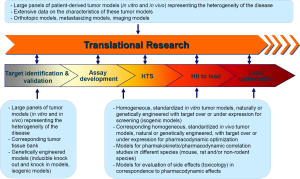
As consequence, primary pharmacology processes in preclinical cancer research have changed (Figure 1). Elementary task is the establishment of the right model and access to appropriate tools for each step of the drug discovery process.
Target identification and validation (TIV) process
Before the introduction of target-specific drug discovery, research was driven primarily by phenotypic screening. However especially in cancer research, the limited knowledge of the molecular mechanisms of disease turned out to be a major disadvantage of the phenotypic screening. The introduction of new technologies to identify targets either in a high throughput setting (i.e., synthetic lethal screens with RNA interference) or by new sequencing techniques, allowing the identification of low frequency disease relevant genetic aberrations, resulted in a tremendous progress and the identification of large numbers of potential targets.
These target-focused approaches provide a specific biological hypothesis which can also be defined as molecular mechanism of action (1). The current challenge is the validation of the hypothesis, especially demonstrating that the specific molecular mechanism is relevant to the disease pathogenesis in a certain population and has a sufficient therapeutic index in the context of the physiological response.
These changes in TIV have also changed the request on the disease models. Have been a handful extensively characterized tumor cell cultures and mouse models been the standard for many decades, the target driven approaches now require models reflecting better the clinical situation (Figure 2).
The requirements on new models include among others:
- large panels of tumor models (in vitro and in vivo) representing the heterogeneity of the disease;
- extensive data about the characteristics of these tumor models (gene and protein expression, gene amplifications, mutations, epigenetics, miRNA expression, histology, reference drug sensitivity);
- corresponding databases containing all these informations and tools allowing bioinformatic analyses;
- tumor tissue banks (frozen and paraffin embedded tissue, tissue micro arrays);
- genetically engineered models (inducible knock out and knock in models, isogenic models).
The target driven drug discovery further requires the definition of strong criteria for the acceptance of the target. The advantage is, that the validation can be supported by first in vivo experiments using molecular and chemical knowledge, applying both small-molecule based strategies (selected compounds from available libraries) and biologicals based approaches, such as individually engineered antibodies.
An important part of the preclinical target validation, next to the molecular mechanism of action, is to investigate possible resistance mechanisms, predictors of response, the identification of rational targets for combinations, and further to analyze the physiological mechanism of action.
As one example, we employed the RNAi screening technology, to determine the modifying effects of reduced gene expression on drug activity (2).
To analyze the mechanisms of mitotic arrest induced by targeting microtubules with a new type of microtubule stabilizer (MTS) and to identify additional targets and biomarkers, a siRNA-based RNAi drug modifier screen was performed in four cancer cell lines. The knockdown of more than 300 genes (900 siRNAs) implicated in cell cycle control, apoptosis, chromosomal instability and taxane-resistance was combined with MTS treatment in a high-throughput RNAi drug modifier screen in three breast cancer cell lines MCF7, T47D and MDA-MB435s and, for comparison, the A549 lung cancer cell line.
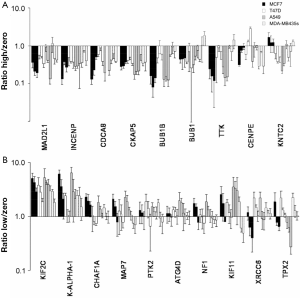
Defects of the spindle assembly checkpoint (SAC) were identified to cause resistance against drug-induced mitotic arrest and apoptosis. The strongest suppressor effects were observed for the knockdown of components of the SAC (Figure 3A). Knockdown of BUB1B, BUB1 and TTK (MPS1) components of the mitotic checkpoint complex, reduced mitotic arrest in MCF7 and A549 cells but had little or no effect on T47D and MDA-MB435s cells. Potential biomarkers for resistance are SAC-defects like mutations in the central SAC-kinase BUB1B.
Chromosomal heterogeneity and polyploidy are also potential biomarkers of resistance since they imply an increased tolerance for aberrant mitosis. RNAi screening showed yet again that the drug is not a substrate of ABC-transporters (2).
The RNAi drug modifier screen demonstrated that the drug-induced mitotic arrest can be enhanced by concomitant inhibition of mitotic kinesins, thus suggesting a potential combination therapy with a KIF2C (MCAK) kinesin inhibitor (Figure 3B). However, the combination of the drug and inhibition of the prophase kinesin KIF11 (Eg5) is antagonistic, indicating that the kinesin inhibitor has to be highly specific to bring about the required therapeutic benefit.
Screening results have been validated in single experiments confirming, that the knockdown of BUB1B or CENPE reduced MTS-induced mitotic arrest in all four cell lines whereas KIF2C knockdown enhanced MTS-induced mitotic arrest. In contrast, a significant reduction of MTS-induced aneuploidy without concomitant increase in G2/M-arrest was seen for KIF11 knockdown in all four cell lines.
To estimate cell survival, a survival index was calculated as the ratio of remaining cell number after MTS treatment divided by initial number of cells numbers. Survival indices were found to be increased for BUB1B knockdown in all four cell lines and for CENPE knockdown in T47D and SKBR3 but decreased for KIF2C knockdown in MCF7 and A549 (2).
As one example how available small molecules can be involved in the target validation, we have elucidated the influence of KIF11 on the induction of aneuploid cells after MTS treatment by comparing the RNAi-mediated knockdown of KIF11 with the effect of ispinesib treatment, a small molecule inhibitor of KIF11 (3). Similar to the RNAi knockdown of KIF11, ispinesib significantly reduced the MTS-induced aneuploidy without increasing mitotic arrest (Figure 4A). The combination of MTS and ispinesib had antagonistic effects in proliferation assays (Figure 4B). Both KIF11 knockdown and KIF11 inhibition caused typical monoasters (Figure 4C,D). Thus, interference with spindle assembly by KIF11 inhibition specifically antagonizes the MTS-induced aneuploidy but not the MTS-induced mitotic arrest.

To conclude, 1 out of the 300 RNAi-targeted genes had a sensitizing effect on MTS in all four cell lines in the screen, and 6 out of the 300 RNAi-targeted genes had a sensitizing effect on MTS in at least two cell lines. On the other hand, 5 out of the 300 RNAi-targeted genes had an antagonistic effect on MTS in all four cell lines in the screen, and eleven out of the 300 RNAi-targeted genes had an antagonistic effect on MTS in at least two cell lines. Validation studies were able to confirm modifier effects for four genes. The study also strongly demonstrates that a panel of heterogenous cell lines needs to be included in these types of assays, as results can be diametral from one cell line to another.
Lead identification and optimization (LO) process
The LO is more or less identical with the classical drug development process. The process will be adapted on the validated targets and includes assay and model development, followed by a screening phase of selected compound, peptide, antibody, or RNAi libraries to identify a lead structure (Figure 1). Once a lead structure has been identified, optimization processes are started, frequently in parallel for several leads.
As the most difficult part of the targeted drug development, this part can be seen as an extended lead and target discovery phase, addressing the molecular mechanism of action in correlation to optimal pharmacodynamic activity (physiological mechanism of action), optimal pharmacokinetics (PK) [absorption-distribution-metabolism-excretion (ADME)], toxicity, as well as resistance development.
A large number of functions are now involved in this integrated preclinical drug development (IPDD, Figure 5), including functions like medicinal and protein chemistry, cell and structural biology, pharmacology, PK and early toxicology (Tox). Data from the screening, now implemented in large data bases, will be further used for computational modelling.

A broad panel of lead optimization tasks and criteria for oncology drug development has been established, which should address:
Predictive pharmacology:
- Demonstrate the extent of target inhibition in correlation to pharmacological effects (i.e., inhibition of tumor growth, -blood flow, -metabolism);
- Identification of main indications [primary tumors, metastases (Mets)];
- Biomarker identification & validation with preclinical models (i.e., by comparison of gene expression profiles from primary tumors);
- Drug sensitivity modifiers screen [i.e., high-throughput screen (HTS) proliferation assays or siRNA technology];
- Combination studies in tumor models;
Resistance:
- Target of drug transporters (ABC transporters), cellular uptake and intracellular distribution;
- Gene regulation by the drug in sensitive and resistant models;
- Mechanisms of apoptosis, mitotic catastrophe and immunomodulation;
Toxicity/PK/imaging:
- Modulation of adverse effects;
- Questions of PK/pharmacodynamics (PD) modeling, scheduling;
- Imaging of response;
Similar to the TIV process, increased demands on the lead optimization have changed the requests on the disease models. The target driven approaches now require models with defined levels of target expression which will be mainly generated by genetic modifications and cloning:
- Homogeneous, standardized in vitro tumor models, naturally or genetically engineered with target over- or under-expression for screening (isogenic models), models for classical drug resistance;
- Homogeneous, standardized in vivo tumor models, natural or genetically engineered with target over- or under-expression for pharmacodynamic optimization (transgenic mice);
- Models for pharmakokinetic/pharmacodynamic correlation studies in different species (mouse, rat and/or non-rodent species) models for evaluation of side effects (Tox) in correspondence to pharmacodynamic effects.
For example, several studies, performed during the development of the already mentioned new MTS, will be discussed. Microtubules are considered as important target for cancer treatments because disruption of microtubule dynamics interferes with cell functions and mitosis, leading ultimately to a G2/M arrest and apoptosis, and several microtubule stabilizing taxane derivatives have been developed as anti-cancer drugs (5). To overcome limitations associated with the established drugs, compounds from different structural classes have been synthesized and tested for activity (6). Extensive preclinical in vitro studies have been set up to demonstrate improved target activity for these new compounds (7).
A defined panel of tumor cell lines (sensitive and multi-drug resistant) was tested in comparison to the available standard (paclitaxel) and found to be strongly sensitive to the new MTS with only moderately variations in response (IC50 between 0.3 and 5.5 nM) (7). So far, no natural resistant cell line was identified and even treatment for more than one year with the new MTS did not result in development of resistance (unpublished own results).
Further mechanistical investigations in tumor cell lines demonstrated, that the new MTS induces a more rapid and potent tubulin polymerization than paclitaxel. A rapid and effective influx into cells, combined with the evasion of P-glycoprotein efflux pumps, have been identified as key qualities resulting in consistently more potent activity than microtubule-stabilizing taxanes (8). However, in line with other MTSs, it causes mitotic arrest, followed by activation of the mitochondrial apoptotic pathway. Profiling of the pro-apoptotic signal transduction pathway using a panel of small interfering RNAs revealed that it acts in a fashion comparable to paclitaxel. In HCT-116 colon cancer cells, the MTS induced apoptosis was partially antagonized by the knockdown of pro-apoptotic members of the Bcl-2 family, including Bax, Bak and Puma, whereas knockdown of Bcl-2, Bcl-XL or Chk1 sensitized cells to cell death (8).
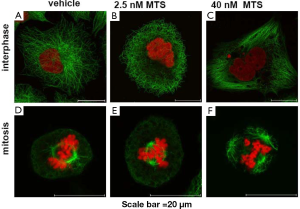
Further mechanistic studies in lung cancer cells (9) revealed a concentration-dependent disturbance of cellular organization with two apparent phenotypes. At low concentrations, an aneuploid phenotype occurred, whereas the classical “mitotic arrest” phenotype was induced only at higher concentrations (Figure 6). Interestingly, the treatment with low doses effectively inhibited cell proliferation, but—compared to high concentrations—induced apoptosis only marginally. Analysis of differential gene expression in tumor cells treated either with high and low drug concentration demonstrated a non-overlapping set of regulated genes:
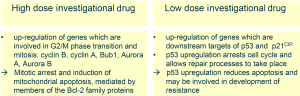
Genes involved in G2/M phase transition and the SAC, like cyclin B1 and bub1b were up-regulated by treatment with high dose MTS. In contrast, treatment with the low concentration revealed an up-regulation of direct transcriptional target genes of TP53, like cdkn1a, mdm2, gadd45a and fas. This resembles an activation pattern which is caused in response to mild, repairable damage, and induces cell cycle arrest, rather than strong damages which promote apoptosis. This allows repair processes to take place and the cells to survive. Knockdown of TP53 led to a significant increase in apoptosis induction (9).
These mechanistic data confirmed, that up-regulation of TP53 and its downstream effectors by low concentrations of MTS is responsible for the relative apoptosis resistance of A549 lung cancer cells and might represent a new mechanism of resistance (Figure 7).
A different phenotype appears to be induced at higher MTS concentrations, with progressively more perturbed microtubule dynamics, formation of microtubule bundles and activation of the SAC leading to an arrest in mitosis. Mainly, this result in an induction of mitochondrial apoptosis, mediated by members of the Bcl-2 family proteins, and is substantially similar to that seen with paclitaxel and other epothilones (8). But, mitotically arrested cells may also undergo aberrant mitosis or mitotic slippage and endo-reduplication. The variations in the extent of apoptosis among breast cancer cells after MTS treatment could be explained by differences in the apoptotic signalling rather than by differences in mitotic arrest.
Translational research (TR) process
TR in oncology from the perspective of the drug developer should provide the simple answer: “who is the right patient for my new drug”, whereas the oncologist is interested in: “which is the right drug for my patient”. This means that in the later stages of cancer drug development and in the management of patients with cancer, “predictive biomarkers” are urgently needed which can be used to identify optimal target populations of patients; predict the efficacy of the drug and patient’s response, resistance and toxicity; and rapidly distinguish between non-responders and patients who respond to therapeutic intervention (10).
The U.S. Food and Drug Administration (FDA)’s Center for Drug Evaluation and Research (CDER) has provided a guidance document on the qualification process for biomarker (titled “Draft Guidance for Industry: Qualification Process for Drug Development Tools”). Requirements set in this document make clear, that the qualification process for a biomarker has many parallels to drug discovery and development, starting with biomarker identification and validation, followed by assay development and optimization, and finally followed by validation in clinical trials. In the preclinical oncology research departments from most pharmaceutical and biotech companies, the TR has now become an integrative part of the development. Considering the heterogeneity of cancer, it has become clear that this research requires new approaches.
As TR needs:
- large panels of patient-derived tumor models (in vitro and in vivo) representing the heterogeneity of the disease;
- extensive data on the characteristics of these tumor models (gene and protein expression, gene amplifications, mutations, epigenetics, miRNA expression, histology, reference drug sensitivity, and corresponding databases containing all this information and tools allowing bioinformatic analyses);
- orthotopic models, metastasizing models, imaging models.
This type of research is now frequently performed in academia-industry partnership.
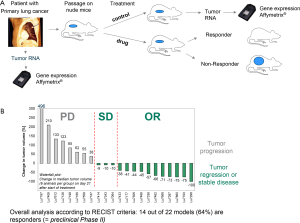
During the development of our previously mentioned MTS, we have addressed the questions for a predictive biomarker in lung cancer patients with a new type of preclinical study. This was based on the observation, that interestingly, some tumor models, i.e., the NCI-H460 lung cancer cells, which are highly sensitive to MTS in cell culture, developed treatment resistant tumors on nude mice (unpublished own results). Human tumors accumulate genetic and molecular abnormalities, leading to broad heterogeneity. Large panels of tumor models reflecting tumor heterogeneity might have increased value for predicting the response to new therapeutic agents in the clinic. Consequently, it is important to use a large panel of clinically relevant tumor models for translational studies. However, from the in vitro studies with 20 breast cancer cell lines and in more than 30 other cell lines, we have not been able to identify natural resistance mechanisms to MTS. This led us to work with extended panels of in vivo models.
To address this discrepancy between in vitro and in vivo activity, further studies across a panel of human lung cancer xenograft models were performed (Figure 8A). In this heterogeneous panel response to MTS—treatment was determined in an integrative preclinical phase II design—further resistant tumors were identified (Figure 8B). We have observed 64% overall responses [response analysis according to the response evaluation criteria in solid tumors (RECIST) clinical trial criteria] with MTS in the 22 non-small cell lung cancer (NSCLC) xenograft models (11). Genome-wide gene expression and mutational analysis were used to identify predictive markers for response and to explore the mechanism of MTS’s anti-tumor activity in vivo. Tumors with wild-type TP53 as well as high expression of genes involved in cell adhesion, hypoxia or angiogenesis were more likely to be resistant to MTS treatment (11). For validation, combination experiments were performed with drugs or siRNA is, targeting some of the identified resistance mechanisms, i.e., tumor angiogenesis, hypoxia or TP53. Indeed, when combined with MTS treatment, combination therapy resulted in restored anti-tumor activity in resistant tumor models [(9,11), unpublished own results)].
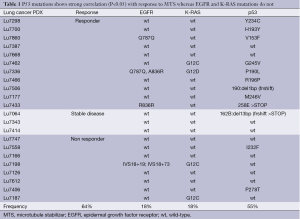
Hypoxia triggers pathways that drive angiogenesis and tumor progression, and the presence of genes associated with these pathways has previously been associated with a negative prognosis and resistance to therapy (12). Up-regulation of CA9 and CA12 gene expression, in particular, has been detected in a large number of common malignancies and is implicated in tumor development (13). The data presented in the NSCLC study show that the combination of MTS with an inhibitor of angiogenesis such as bevacizumab or sorafenib results in an enhanced antitumor effect in tumor models with an activated HIF1a/hypoxia pathway (11). No correlations were found between MTS activity and overexpression or mutations of egfr and k-ras genes suggesting that MTS may be active in patients with NSCLC tumors with these changes (Table 1).
Full table
In our NSCLC xenograft study, response to MTS correlated with low expression or expression of mutant TP53 (Table 1). In cell culture studies, we performed earlier, treatment of A549 cells with low concentrations of MTS resulted in stabilization of TP53 and induction of TP53 target genes, potentially resulting from consistent translation of the long-lived TP53 mRNA during prolonged mitosis (14). TP53 check point induction by low MTS concentrations targets genes such as cdkn1a or gadd45a and induces cell cycle G1 arrest, rather than promoting apoptosis (15-17). This may allow for repair processes cell survival. It might be possible that in tumors, harboring areas with low vascularization, only very low amounts of MTS will actually reach the tumor cells. In terms of chemotherapy, this would indicate an unfavorable condition, because cells might start re-growing after the cell cycle arrest. In vitro, as we have demonstrated here, the MTS-induced aneuploid cells may arrest permanently or enter senescence. Yet, it is an open question whether in vivo these cells undergo apoptosis, enter senescence or start re-growing eventually. Previously, we have shown that the knockdown of TP53 increased the rate of apoptosis after MTS treatment in A549 lung cancer cells (9). Additionally, in our studies on patient- derived NSCLC xenografts, a pronounced long-term response to MTS was seen when TP53 was mutated. The question remains whether these tumors might have a higher probability to respond to MTS, therefore investigations, whether mutational status of TP53 could serve as predictive biomarker in clinical trials, warrants further investigation. Additionally, it could be of clinical relevance if patients with TP53 wild type tumors benefit from combination therapy with drugs inhibiting TP53 or only certain specific functions of TP53, i.e., blocking TP53-dependent transactivation with no effect on p53-mediated apoptosis.
In conclusion, results have been generated from a large set of patient-derived xenograft models via genome-wide gene expression analysis, and mutation analysis of selected genes to identify potential markers of response and refractoriness to MTS in NSCLC. Our data suggest that MTS may be active where other chemotherapies are not. Clinical investigations of the marker genes (e.g., CA9, CA12, EPHA4, ITGA6) together with TP53 gene expression and mutation analysis could be used as predictive marker.
Besides these mechanistic molecular biology driven studies, more classical pharmacology studies have been performed to demonstrate effects of MTS on brain and bone metastases. Taxanes are unable to cross an intact blood-brain barrier, which can result in the lack of activity against brain metastases (18). We investigated the activity of MTS in new models for brain metastasis of breast and lung cancer, respectively.
Our studies aimed to determine whether MTS could cross the blood-brain barrier and reduce brain tumor/metastases growth more effectively than other anticancer agents in clinically relevant human tumor models (19). The preclinical studies provided direct evidence that MTS has free access to the brain, leading to highly effective levels of the drug in the brain tissue, which maintained for several days. In vivo studies demonstrated that MTS resulted in significant inhibition of tumor growth in both the subcutaneous and intracerebral glioblastoma xenograft models, whereas paclitaxel showed consistent activity in the subcutaneous models only. Similarly, in models of brain metastases, including patient-derived models of NSCLC, MTS showed superior antitumor activity against brain tumors compared with paclitaxel or temozolomide (19).
Bones are a preferred site for metastases in patients with breast cancers. We showed that MTS inhibited tumor burden and bone destruction, in addition to reducing tumor-induced cachexia and paraplegia. MTS treatment significantly lowered the number of activated osteoclasts and significantly reduced the osteolytic lesion area, bone volume loss, and bone resorption, inhibiting the vicious cycle of both tumor growth and bone resorption, suggesting a substantial benefit in the treatment of patients with breast cancer at risk from bone metastases (20).
Summary and outlook
What have been the “lessons learned” from the preclinical development of MTS? Depending on the stage of the drug discovery program, different models are required. For primary in vitro screening, cell lines can be utilized easily from the available large panels or generated by genetic engineering. They can be selected based on the target or the question to be answered. For example, we have used a pair of cell lines with high and low P-glycoprotein expression to optimize our MTS against drug efflux pumps causing multidrug resistance (7). For secondary in vitro screening, larger panels of tumor cell lines with known sensitivity or resistance to available standard drugs are used for further profiling.
However, as we have learned from our mechanistic studies with HCT-116 cells (8), A549 cells (9) and from the drug sensitivity modifier screen reported here using MCF7, T47D, A549 and MDA-MB435s cells, it is of utmost importance to perform these studies in a panel of three or more different tumor models. If we have performed the RNAi drug modifier screen in only one cell line, we would on the one hand have missed important targets which we have seen only in the other three cell lines (e.g., KIF11, CENPE), and on the other hand, we would have identified many modifying genes which turned out to be not relevant in other cell lines. The in vitro mechanistic studies revealed rather general mechanisms involved in apoptosis induction (Bcl-2 family and Bax) or cell cycle arrest (tumor suppressor TP53 or SAC kinases) to be involved in the sensitivity to MTS. However, the identification of KIF2C (MCAK) knockdown, synergizing with MTS effects, has impressively shown the potential of this technology. Thus, KIF2C inhibition seems to be a valuable combination strategy for MTS.
Looking at in vivo anti-tumor models, a differential pattern of sensitivity can be observed. Broad activity was also seen in most of these models, however most interestingly, some tumor models, i.e., the NCI-H460 lung cancer cells, which are highly sensitive to MTS in cell culture, developed treatment resistant tumors on nude mice. To address this gap between in vitro and in vivo activity, further studies need to be performed. This gap also reminds us that in vivo experiments are still crucial and remain an integral part to evaluate tumor response in the near future.
Although mouse xenograft models derived from established human cancer cell lines have undoubtedly enhanced the understanding of the anti-tumor activity of novel anti-cancer agents, these models have several disadvantages. Depending on the number of cell passages, xenografts can behave very differently to the primary tumor (21), and combined with other deficiencies in pre-clinical approaches [reviewed in (22)], this can reduce the relevance of established xenograft models for predicting the probability of success of anti-cancer drugs in clinical studies for some tumor localizations. Analysis of antitumor activity in patient-derived xenograft models has provided a more accurate selection process for the identification of agents which have activity in clinical trials, suggesting that some of these models may provide a useful hint for activity in the clinic (23). Genome-wide analyses of gene expression using oligonucleotide microarrays have allowed the determination of molecular characteristics present in xenograft models that mirror tumor behavior and relate to disease progression and survival (24). Furthermore, correlations between the growth of xenograft models derived directly from patient tumors and the clinical prognosis of donor patients have been reported (25,26). In the future, the use of patient-derived human tumor xenografts will therefore play a key role in the search for more efficacious cancer treatments (27-31). The ability to identify and assess anti-tumor activity in well-characterized xenografts in correlation with particular genetic or molecular characteristics may aid the development of new therapeutic regimens. In our studies, increased basal expression of genes involved in cell adhesion, angiogenesis and the hypoxia pathway was observed in lung cancer xenograft models that do not respond to MTS. In these models, the combination of MTS with drugs targeting VEGF signaling led to an enhanced anti-tumor activity compared with either agent alone.
Conclusions from what we discussed here are:
- Drug discovery, systems biology, and TR are moving together to address all the new hallmarks of cancer increasing the success rate of drug discovery;
- In vitro versus in vivo models or vice versa—as we have shown both models have limitations and advantages, however, when used critically, all generate important and reliable results;
- Panels of patient derived xenograft models represent an important tool for TR;
- Predictive value of the preclinical models is increasing steadily, however, even genetically engineered “humanized” mice are still not men.
Acknowledgements
The author would like to thank S. Winsel, J. Eschenbrenner, K. Mittelstaedt, M. Drosch, A. Sommer, U. Klar, S. Hammer, B. Weiss, C. Merz, G. Siemeister, S. Käkönnen, and A. Strube from Bayer Pharma AG who have performed many of the experiments described in the review.
Further I would like to thank many cooperation partners who significantly contributed to the TR reviewed in the paper: C. Sachse, M. Hannus and M. Seidel from Cenix BioScience GmbH Dresden, I. Vitale and G. Kroemer from IGR Paris, and I. Fichtner, J. Rolff, M. Becker, M. Lemm from EPO Berlin-Buch GmbH.
I would also like thank K. Bosslet and D. Mumberg for their strong scientific support developing TR programs.
Disclosure: J. Hoffmann is former employee of Bayer Pharma AG and holds patents on the compounds as well as shares of Bayer AG.
References
- Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov 2011;10:507-19. [PubMed]
- Eschenbrenner J, Winsel S, Hammer S, et al. Evaluation of activity and combination strategies with the microtubule-targeting drug sagopilone in breast cancer cell lines. Front Oncol 2011;1:44. [PubMed]
- Purcell JW, Davis J, Reddy M, et al. Activity of the kinesin spindle protein inhibitor ispinesib (SB-715992) in models of breast cancer. Clin Cancer Res 2010;16:566-76. [PubMed]
- Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 2006;58:621-81. [PubMed]
- Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253-65. [PubMed]
- Klar U, Hoffmann J, Giurescu M. Sagopilone (ZK-EPO): from a natural product to a fully synthetic clinical development candidate. Expert Opin Investig Drugs 2008;17:1735-48. [PubMed]
- Klar U, Buchmann B, Schwede W, et al. Total synthesis and antitumor activity of ZK-EPO: the first fully synthetic epothilone in clinical development. Angew Chem Int Ed Engl 2006;45:7942-8. [PubMed]
- Hoffmann J, Vitale I, Buchmann B, et al. Improved cellular pharmacokinetics and pharmacodynamics underlie the wide anticancer activity of sagopilone. Cancer Res 2008;68:5301-8. [PubMed]
- Winsel S, Sommer A, Eschenbrenner J, et al. Molecular mode of action and role of TP53 in the sensitivity to the novel epothilone sagopilone (ZK-EPO) in A549 non-small cell lung cancer cells. PLoS One 2011;6:e19273. [PubMed]
- Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov 2012;11:201-14. [PubMed]
- Hammer S, Sommer A, Fichtner I, et al. Comparative profiling of the novel epothilone, sagopilone, in xenografts derived from primary non-small cell lung cancer. Clin Cancer Res 2010;16:1452-65. [PubMed]
- Cosse JP, Michiels C. Tumour hypoxia affects the responsiveness of cancer cells to chemotherapy and promotes cancer progression. Anticancer Agents Med Chem 2008;8:790-7. [PubMed]
- Brahimi-Horn MC, Chiche J, Pouysségur J. Hypoxia and cancer. J Mol Med (Berl) 2007;85:1301-7. [PubMed]
- Blagosklonny MV. Mitotic arrest and cell fate: why and how mitotic inhibition of transcription drives mutually exclusive events. Cell Cycle 2007;6:70-4. [PubMed]
- Aylon Y, Oren M. Living with p53, dying of p53. Cell 2007;130:597-600. [PubMed]
- Riley KJ, Maher LJ 3rd. p53 RNA interactions: new clues in an old mystery. RNA 2007;13:1825-33. [PubMed]
- Das S, Boswell SA, Aaronson SA, et al. P53 promoter selection: choosing between life and death. Cell Cycle 2008;7:154-7. [PubMed]
- Kastritis E, Efstathiou E, Gika D, et al. Brain metastases as isolated site of relapse in patients with epithelial ovarian cancer previously treated with platinum and paclitaxel-based chemotherapy. Int J Gynecol Cancer 2006;16:994-9. [PubMed]
- Hoffmann J, Fichtner I, Lemm M, et al. Sagopilone crosses the blood-brain barrier in vivo to inhibit brain tumor growth and metastases. Neuro Oncol 2009;11:158-66. [PubMed]
- Strube A, Hoffmann J, Stepina E, et al. Sagopilone inhibits breast cancer bone metastasis and bone destruction due to simultaneous inhibition of both tumor growth and bone resorption. Clin Cancer Res 2009;15:3751-9. [PubMed]
- Haddad TC, Yee D. Of mice and (wo)men: is this any way to test a new drug? J Clin Oncol 2008;26:830-2. [PubMed]
- Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov 2006;5:741-54. [PubMed]
- Furman WL, Stewart CF, Poquette CA, et al. Direct translation of a protracted irinotecan schedule from a xenograft model to a phase I trial in children. J Clin Oncol 1999;17:1815-24. [PubMed]
- Nevins JR, Huang ES, Dressman H, et al. Towards integrated clinico-genomic models for personalized medicine: combining gene expression signatures and clinical factors in breast cancer outcomes prediction. Hum Mol Genet 2003;12:R153-7. [PubMed]
- Angevin E, Glukhova L, Pavon C, et al. Human renal cell carcinoma xenografts in SCID mice: tumorigenicity correlates with a poor clinical prognosis. Lab Invest 1999;79:879-88. [PubMed]
- Peterson JK, Houghton PJ. Integrating pharmacology and in vivo cancer models in preclinical and clinical drug development. Eur J Cancer 2004;40:837-44. [PubMed]
- Perez-Soler R, Kemp B, Wu QP, et al. Response and determinants of sensitivity to paclitaxel in human non-small cell lung cancer tumors heterotransplanted in nude mice. Clin Cancer Res 2000;6:4932-8. [PubMed]
- Fichtner I, Becker M, Zeisig R, et al. In vivo models for endocrine-dependent breast carcinomas: special considerations of clinical relevance. Eur J Cancer 2004;40:845-51. [PubMed]
- Becker M, Sommer A, Krätzschmar JR, et al. Distinct gene expression patterns in a tamoxifen-sensitive human mammary carcinoma xenograft and its tamoxifen-resistant subline MaCa 3366/TAM. Mol Cancer Ther 2005;4:151-68. [PubMed]
- Fichtner I, Rolff J, Soong R, et al. Establishment of patient-derived non-small cell lung cancer xenografts as models for the identification of predictive biomarkers. Clin Cancer Res 2008;14:6456-68. [PubMed]
- Garber K. From human to mouse and back: ‘tumorgraft’ models surge in popularity. J Natl Cancer Inst 2009;101:6-8. [PubMed]