Genotyping of cutaneous melanoma
Introduction
Melanoma has the highest mutation rate of all common cancers (1,2). Over time, this discovery has led to a dramatic increase in the understanding of the molecular features, drivers, and heterogeneity of this disease. The management of melanoma is now evolving rapidly due to the expanded availability and use of high-throughput sequencing technologies that provide broad genetic assessment of tumors (3). The identification of the many somatic events that occur in melanomas has been complemented by functional studies which have helped to illuminate the critical pathways and processes affected by these changes. Most importantly, research has validated the clinical significance of many of these genetic events, both as prognostic markers and as therapeutic targets (4).
While the field of melanoma has been advanced significantly by genomic research, this new information also comes with challenges (5). Physicians are now faced with a growing number of options to genotype melanoma patients (6). Based on these tests, and increasingly by testing requested and/or procured by patients themselves, physicians are being asked about the clinical implications of a growing number of molecular alterations. This review will summarize the current understanding of the genetic landscape of mutations in melanoma, their associations with clinicopathological features, and their implications for testing and treatment.
The RAS-RAF-MEK-ERK pathway
The RAS-RAF-MEK-ERK signaling pathway is an important regulator of cellular proliferation and survival that has been implicated in multiple tumors types (7). Initial focused studies, and more recently whole exome sequencing, support that this pathway is affected by activating events more frequently than any other pathway in cutaneous melanomas (8,9). A number of these mutations have been shown to correlate with patient and tumor features. Most importantly, several of the mutated genes have been validated in patients as therapeutic targets.
BRAFV600 mutations
Like the other members of the RAF family of kinases (ARAF, CRAF), BRAF is a serine/threonine protein kinase. One of the sentinel events in understanding the molecular pathogenesis of melanoma was the discovery of mutations in the BRAF gene that result in substitutions at the V600 residue of the protein (BRAFV600 mutations). In the initial cohort examined, BRAFV600 mutations were identified in ~15% of tumors from various human cancers, and strikingly in ~60% of melanomas (10). Subsequent studies have demonstrated that BRAFV600 mutations are present in almost 80% of human melanoma cell lines derived from cutaneous melanomas (11). Large single-center studies and meta-analyses have reported BRAFV600 mutation rates of 40-45% in clinical specimens (11-13). This difference between clinical samples and cell lines likely reflects a selective advantage for in vitro growth and survival for melanomas with BRAFV600 mutations. There are a number of different BRAFV600 mutations observed. The most common mutation results in the substitution of valine with glutamic acid (BRAFV600E), which represents ~70% of detected BRAF mutations (12,14). Mutations that result in substitution with lysine (BRAFV600K) are the second most common (~20%), while other rare substitutions include BRAFV600Dand BRAFV600R. In vitro studies demonstrated that all of the BRAFV600 mutations result in markedly increased kinase activity of the BRAF protein (>200-500 fold) and lead to constitutive activation of downstream components of the RAS-RAF-MEK-ERK pathway (15,16).
The presence of BRAFV600 mutations is significantly associated with both molecular and clinical features. BRAFV600 mutations and mutations in NRAS, which are detected in ~20% of cutaneous melanomas and also activate the RAS-RAF-MEK-ERK pathway, are essentially mutually exclusive in treatment-naïve patients (17,18). In one study of 677 melanomas with molecular testing for BRAFV600 and NRAS mutations, only four tumors (0.6%) had both mutations present (12). BRAFV600 mutations are present in 40-50% of cutaneous melanomas arising in areas intermittently exposed to the sun (19). However, their prevalence is lower in cutaneous melanomas that occur on skin that is chronically exposed to the sun (5-20%) (20-22). BRAFV600 mutations are also relatively rare (10-15%) in acral melanomas, which occur on palms, soles, or nail beds, and in mucosal melanomas (~5%), and are virtually never found in uveal melanomas (23-25). Interestingly, BRAFV600 mutations are also found in up to 82% of benign nevi (26,27). Consistent with this likely early role in tumor development, studies in which BRAFV600 mutation status has been examined in multiple tumors from individual patients have demonstrated concordance rates of ≥90% (28).
Retrospective analyses of cutaneous melanomas have generally shown that the presence of a BRAFV600 mutation is associated with younger age at diagnosis, a lack of evidence of chronic sun-damage, and superficial spreading or nodular histology in the antecedent primary melanoma (29,30). BRAFV600 mutations are not significantly associated with shorter time to distant metastasis or overall survival (OS) from primary tumor diagnosis (31,32). Two studies did identify significant associations with OS from stage IV. In one study in which only the BRAFV600 mutation status was determined, patients with a mutation had significantly shorter OS from stage IV compared to all patients without a BRAFV600 mutation (30). In another study in which NRAS mutations were also assessed, the presence of a BRAFV600 mutation was associated with shorter OS after stage IV diagnosis compared to patients who had neither BRAF nor NRAS mutations (29). However, the OS of patients with NRAS and BRAFV600 mutations were very similar. More recently, these cohorts of stage IV patients have been interrogated for significant associations with specific BRAFV600 mutations. Both analyses demonstrated that metastatic melanoma patients with BRAFV600K mutations were older at diagnosis and were more likely to have a primary tumor that had evidence of, or arose in areas associated with, chronic sun damage (CSD) compared to patients with BRAFV600E mutations (33,34). The presence of a BRAFV600K mutation was also associated with shorter time from initial diagnosis to stage IV disease, and shorter OS after stage IV diagnosis, compared to BRAFV600E. While these findings are intriguing, these associations need to be evaluated in series of primary melanomas that include patients who did not go on to develop stage IV disease to fully understand their clinical associations and prognostic significance.
Three targeted therapies have been approved to date for the treatment of patients with stage IV or unresectable melanoma with BRAFV600 mutations. Vemurafenib and dabrafenib are potent and selective small molecule inhibitors of BRAFV600 mutant proteins. Preclinical studies demonstrated that both of these agents blocked growth, survival, and MAPK pathway activation in vitro and in vivo in human melanoma cell lines with BRAFV600 mutations (35-37). In contrast, both agents cause activation of the MAPK pathway and they increased growth in vitro and in vivo when they were tested in cell lines with a wild-type BRAF gene (38-40). Thus, testing for the presence of a BRAFV600 mutation is essential for any patient in which these agents are considered. Vemurafenib and dabrafenib both achieved significant improvements in overall response rates (ORR, ~50%), disease control rate (DCR, ~90%), and progression free survival (PFS, median ~6 months) compared to chemotherapy in randomized phase III trials in metastatic melanoma patients with BRAFV600 mutations (41,42). While most patients in these clinical trials had BRAFV600E mutations, the clinical testing of dabrafenib also included patients with known BRAFV600K mutations. Although the patients with BRAFV600K mutations gained clinical benefit from treatment with the selective BRAF inhibitor, the ORR and PFS in these patients was lower than was observed in the patients with BRAFV600E mutations (43-45). Other small case series have reported potentially increased efficacy of BRAF inhibition in patients with BRAFV600R mutations (46-48). Trametinib, the third oral targeted therapy approved for the treatment of metastatic melanoma patients with BRAFV600 mutations, is a selective inhibitor of MEK1 and MEK2. The ORR (22%), PFS (median 4.8 months), and OS [hazard ratio (HR) 0.54] observed with trametinib were superior to chemotherapy in a randomized phase III trial (49). While these outcomes appear to be less impressive than those achieved with BRAFV600 inhibitors, the combination of dabrafenib and trametinib significantly increased response rates (76% vs. 54%) and PFS (median 9.4 vs. 5.8 months), and decreased the rate of cutaneous squamous cell carcinomas (7% vs. 19%), compared to treatment with dabrafenib alone in a randomized phase II trial (50). The combination of dabrafenib and trametinib was approved for the treatment of BRAFV600 mutant metastatic melanoma in 2014.
Other BRAF alterations
To date more than 30 mutations in the BRAF gene have been identified in cancer (51). The majority of these mutations have been identified in exon 15, which includes the region encoding the V600 residue, and in exon 11. In vitro testing of a spectrum of mutations affecting sites in BRAF other than V600 (BRAFNon-V600) demonstrated that the resulting mutant proteins are markedly heterogeneous in their catalytic activity (51). Some of the mutations markedly increase the kinase activity of the BRAF protein, and thus directly lead to increased activation of MEK and ERK. Some BRAFNon-V600 mutations cause very little increase in kinase activity, while others actually decrease the catalytic activity of the BRAF protein. These mutations still appear to cause increased RAS-RAF-MEK-ERK signaling, however, due to increased heterodimer formation by the mutant BRAF proteins with other RAF kinases (i.e., CRAF) (51,52). In contrast to BRAFV600 mutations, it has been noted that melanomas with BRAFNon-V600 mutations often have concurrent activating NRAS mutations (18,29).
Initial in vitro testing of melanoma cells with BRAFNon-V600 mutations demonstrated that such cells were sensitive to growth inhibition by sorafenib, which is a potent inhibitor of CRAF (53). Other studies have shown that melanoma cell lines with BRAFNon-V600 mutations are sensitive to MEK inhibitors (54). Dramatic and durable responses to MEK inhibitors have been reported in two patients with BRAFL597 mutations in early phase clinical trials of the MEK inhibitors TAK-733 and trametinib (54,55). A second patient in the phase I trial of trametinib with a BRAFNon-V600 mutation (BRAFG469A) achieved a minor response (55). A durable (>4 years) complete response to dasatinib treatment has also been reported in a non-small cell lung cancer patient with an inactivating BRAFY472C mutation, with in vitro data mechanistically supporting that inactivating BRAFNon-V600 mutations confer sensitivity to that agent (56).
In addition to mutations, recent studies have identified gene fusions involving BRAF in cancer. BRAF fusions were initially identified as rare events in prostate cancer, pilocytic astrocytomas, gastric adenocarcinomas, and thyroid cancer (57). Examination of 131 melanocytic lesions identified one congenital melanocytic nevus with a BRAF fusion. A subsequent melanoma-specific study identified two additional BRAF fusions. Both were detected in tumors that lacked BRAFV600 and NRAS mutations; overall, 2 of 24 (8%) such “wild-type” tumors harbored BRAF rearrangements. Analysis of the publicly available melanoma TCGA data for 49 tumors with wild-type BRAFV600 and NRAS status identified 2 additional fusions (4.1%) (58). Expression of one of the BRAF fusions resulted in increased activation of the MAPK pathway which was sensitive to trametinib (MEKi), but not vemurafenib (BRAFi). A third study of comparative genome hybridization (CGH) data from 848 melanocytic lesions identified BRAF fusions in ten samples (59). For the six cases with sufficient DNA available for extended analysis, the fusion events were confirmed in each tumor, and no BRAFV600 or NRAS mutations were present concurrently. The fusions did not affect the kinase domain of BRAF, but instead interrupted an inhibitory domain. One fusion event was found in a cell line that showed increased sensitivity to sorafenib and decreased sensitivity to vemurafenib compared to two human melanoma cell lines with BRAFV600 mutations. Interestingly, the patient from whom the cell line was derived had a prolonged clinical response to sorafenib (59).
NRAS
The RAS genes (HRAS, KRAS and NRAS) encode small GTPases that generally trigger the activation of the RAS-RAF-MEK-ERK cascade (60). While activating KRAS mutations have been found in many different cancers, they are extremely rare in melanoma. NRAS mutations are found in about 20% of cutaneous melanomas, 10% of acral melanomas, and 5-13% of mucosal melanomas (21,25,61,62). Similar to BRAFV600 mutations, NRAS mutations are not found in uveal melanomas, but they have been detected in cutaneous nevi (25,62,63). The overwhelming majority of NRAS mutations affect the nucleotides encoding the G12, G13, and Q61 residues of the protein (11,29). While these are identical to the sites most commonly affected in KRAS in other cancers, in melanoma the majority (~80%) of the mutations affect Q61, whereas KRAS mutations generally affect G12/13 (29,64). A recent analysis of 136 advanced-stage melanoma patients with NRAS mutations did not find any significant differences in the patient demographics, primary tumor characteristics, or clinical outcomes between patients with NRAS exon 1 (G12/13) and exon 2 (Q61) mutations (34). As noted previously, NRAS mutations and BRAFV600 mutations are mutually exclusive in newly diagnosed melanomas (18). Thus, molecular testing for NRAS can increase the confidence of a negative clinical test for a BRAFV600 mutation clinically.
Retrospective analyses have reported that the presence of a NRAS mutation is associated with older age at diagnosis, primary tumor location on the extremities, and nodular histology (29,65,66). In contrast to BRAFV600 mutations, the presence of a NRAS hotspot mutation has been associated with shorter time to distant metastasis and shorter OS after initial diagnosis (31). The presence of NRAS mutation was also significantly associated with shorter OS after the diagnosis of stage IV in a cohort of advanced melanoma patients (29). Thus, the development of effective therapeutic strategies for NRAS-mutant melanomas is a high priority and critical unmet need.
Preclinical studies have demonstrated that treatment of human tumor cells with activating RAS mutations with mutant-selective BRAF inhibitors (vemurafenib, dabrafenib) paradoxically accelerates their growth in vitro and in vivo (38-40,67). In contrast, MEK inhibitors have shown some promise in this molecularly defined melanoma subtype. A phase II study of the MEK inhibitor MEK162 in 30 metastatic melanoma patients with activating NRAS mutations reported an ORR of 20% and a DCR of 63% (68). However, the majority of the clinical responses were quite short, and the median PFS was only 3.7 months in these patients. In the phase I trial of trametinib, 0 of 7 melanoma patients with NRAS mutations responded (55). Preclinical studies support that effective targeted therapy for melanomas with NRAS mutations will likely require MAPK pathway inhibitors to be combined with other agents (69,70). Experiments in genetically engineered mouse models (GEMMs) of NRAS-mutant melanoma suggest that combined inhibition of MEK and the cyclin-dependent kinase 4 (CDK4) can produce complete regressions in ~1/3 of these tumors (71,72). Multiple clinical trials testing CDK4 inhibitors as single agent and in combination with MEK inhibitors in NRAS-mutant melanoma have been initiated (NCT01781572, NCT01037790, NCT01820364, www.clinicaltrials.gov) (72).
MEK1/2 mutations
The serine-threonine kinases MEK1 and MEK2 transmit signals downstream from the RAF proteins. Recent broad sequencing studies have identified mutations in MEK1/2 in ~6% of cutaneous melanomas (8,9,63,73). Some of the mutations in MEK1 that have been characterized are highly activating and can mediate resistance to both BRAF and MEK inhibitors (74). However, other mutations have been detected in melanomas with concurrent BRAFV600 mutations in patients that have responded to vemurafenib (75,76). Thus, the functional significance of MEK1/2 mutations is likely heterogeneous, similar to the existing data regarding BRAFNon-V600 mutations. Due to their relative recent discovery and low prevalence, at this time there is very little information about the demographics, tumor features, and clinical outcomes associated with these mutations.
NF1
The NF1 gene encodes neurofibromin 1. NF1 is a RAS-GTPase-activating protein (RAS-GAP) which negatively regulates the activity of the RAS proteins (77). Loss of NF1 causes neurofibromatosis type 1, a familial disorder associated with abnormalities in the nervous system, skin, and bones (78). Whole exome sequencing of 121 melanomas identified loss of function NF1 mutations in 7 tumors (5.7%) (9). While overall this incidence is relatively low, the mutations were detected in 25% of the melanoma with wild-type BRAF and NRAS genes, which was significantly higher than the rate (2%) in tumors with mutations in either of those genes. Thus, NF1 mutations appear to be a common genetic mechanism to activate the MAPK pathway in melanomas without BRAFV600 or NRAS mutations, and thus may define a new molecular subset of this disease. In addition to this suggestive data from sequencing studies, functional studies in a BRAFV600 GEMM demonstrated that loss of NF1 reduces BRAF-induced senescence and enhances melanoma formation (79). This study and another independent report also showed that the loss of NF1 expression can confer resistance to selective BRAF inhibitors in melanomas with BRAFV600 mutations (80).
KIT mutations and amplifications
The c-KIT gene encodes a receptor tyrosine kinase (RTK) that can activate multiple downstream signaling pathways, including the RAS-RAF-MEK-ERK and PI3K-AKT cascades (81). During development, KIT activity supports developing melanocytes and their migration, and several studies demonstrated that melanoma progression is generally associated with loss of KIT expression and function (82,83). Thus, early data suggested that KIT does little to contribute actively to the pathogenesis of melanoma. However, recent studies now support that in fact KIT frequently plays an important role in the pathogenesis of non-cutaneous melanomas.
The first study to demonstrate that acral melanomas, mucosal melanomas, and cutaneous melanomas with CSD have relatively low rates of BRAFV600 and NRAS mutations identified several chromosomal regions with frequent copy number gain in these melanoma subtypes (21,84). Subsequent focused analysis of the 4q12 chromosomal region, which harbors several candidate oncogenes, identified both focal amplifications and somatic mutations in the c-KIT gene (84). These and subsequent studies have gone on to show that somatic mutations in c-KIT are present mainly in mucosal melanoma, but they can also be found in CSD cutaneous and acral, melanomas (22,85-89). Mutations in c-KIT appear to be quite rare (1-2%) in cutaneous melanomas without CSD, but they have been reported. The most common mutations identified in melanoma are involve KITL576P (exon 11) and KITK642E (exon 13), but many other mutations have been identified, often only in individual patients (90). Gene amplification of both wild-type and mutant alleles of c-KIT have also been identified in the same melanoma subtypes in which the mutations are frequently found.
KIT inhibitors are the standard of care in other diseases with frequent c-KIT mutations, such as gastrointestinal stromal tumors (GIST). Notably, the mutations that are detected in melanomas predominantly affect the same exons that are frequently mutated in GISTs. c-KIT mutation status has not been identified as an independent predictor of time to metastases or survival in patients with metastases (91). A number of case reports have demonstrated that metastatic melanoma patients with c-KIT mutations can have dramatic and durable clinical responses to KIT inhibitors (83). The results of three phase II clinical trials of the KIT inhibitor imatinib in patients with c-KIT mutations and/or amplifications have been reported to date. Clinical response rates of 16-30% have been reported in these trials (92-94). These response rates are much higher than those observed in three previous phase II trials that did not include molecular inclusion criteria and predominantly enrolled cutaneous melanoma patients. However, it remains unclear why the response rate has been much lower than is observed in c-KIT-mutant GIST patients. Thus, while molecular testing for c-KIT mutations can identify patients with an increased chance of responding to KIT inhibitors, additional work in ongoing to refine testing strategies to optimize this testing and treatment strategy.
Cell cycle regulators
A significant role in melanoma for genes that regulate cell cycle progression was initially identified based on the molecular characterization of familial melanomas (95). The most common germline aberrations found in such families are loss of function mutations in the CDKN2A gene (96). Through different transcriptional initiation sites the CDKN2A gene encodes two proteins, p16INK4A and P14ARF (72). p14ARF regulates the DNA damage response and apoptosis by inhibiting MDM2, which normally inhibits p53 function. p16INK4A normally functions by binding to CDK4 (97). This interaction inhibits the activity of the CDK4-CyclinD1 complex, which normally promotes cell cycle progression by phosphorylating RB1. Underscoring the importance of this pathway, germline mutations in CDK4 that abolish the site that mediates binding to p16INK4A are the most common germline aberration in families without CDKN2A mutations (98). In addition to these germline events, melanomas have been found to harbor somatic CDKN2A deletions, CDK4 amplifications, and CCND1 amplifications (99). p16INK4A expression and function can also be lost due by methylation of CDKN2A (100,101). Overall, DNA alterations that result in dysregulation of this pathway are detected in over 90% of melanoma cases (72,99). As such, they are detected in melanomas with and without BRAFV600 or NRAS mutations (102).
The alterations of this pathway in familial melanoma strongly supported a role for the pathway in early melanoma development. This hypothesis is also supported by experiments in animal models (103). More recently, analysis of pre-treatment clinical specimens of patients with BRAFV600 mutations who had been treated with dabrafenib found that the presence of low gene copy number of CDKN2A or high copy number of CCND1 were associated with shorter PFS (104). Based on these findings, there is a strong rationale to evaluate agents that target this pathway clinically. A variety of CDK inhibitors are now undergoing clinical testing. Preclinical studies in human melanoma cell lines have shown that the presence of a detectable alteration in CDKN2A, CDK4, or CCND1 correlates with increased sensitivity to single-agent treatment with CDK4 inhibitors (100). As described above, the combination of MEK inhibitors and CDK4 inhibitors are currently being investigated in melanomas with activating NRAS mutations, as are combinations with BRAF inhibitors in melanomas with BRAFV600 mutations. The high prevalence of aberrations in other MAPK pathway genes and cell cycle regulators suggest that this combination may also be rational to explore in patients with BRAF/NRAS-‘wild-type’ melanoma.
The PI3K-AKT pathway
The PI3K-AKT pathway is an important regulator of cell growth, proliferation, differentiation, metabolism, motility, and survival (70,105). Studies have demonstrated that this pathway can be activated genetically multiple ways in cancer (106). However, the prevalence of these alterations varies markedly by tumor type.
While mutations in NRAS and c-KIT may mediate their oncogenic effects at least in part through the PI3K-AKT pathway, hotspot activating mutations in the core pathway components appear to be relatively rare in melanoma. PIK3CA, which encodes the catalytic subunit of PI3K and is frequently affected by driver mutations in breast and colon cancer, is mutated in 2-4% of melanomas, often at sites of unknown functional significance (107). Mutations that cause an E17K substitution in AKT1, which were previously identified as rare activating events in other tumor types, have been identified in ~1% of melanomas (108). Mutations at this same locus in AKT3 have also been identified in ~1% of melanomas. This finding adds to previous studies implicating AKT3 in melanoma progression and metastasis.
PTEN is a lipid phosphatase that dephosphorylates the residue on lipids that is phosphorylated by PI3K. Loss of PTEN causes increased and constitutive activation of the PI3K-AKT pathway (109). PTEN loss of function has been detected in 10-30% of melanomas, due to microdeletions, frameshift mutations, and/or epigenetic mechanisms (110-112). PTEN loss is largely mutually exclusive with NRAS mutations, but it has been detected in BRAFV600 mutated and in BRAF/NRAS-‘wild-type’ melanomas (113-115). The functional and clinical significance of the co-occurrence with BRAFV600 mutations is supported by multiple studies. GEMMs in which the BRAFV600 protein is expressed in melanocytes develop melanocyte hyperplasia but not melanomas. However, concurrent loss of PTEN in that GEMM results in 100% incidence of invasive melanomas that spontaneously metastasize (116). Multiple studies have shown that loss of PTEN also reduces the sensitivity of human melanoma cell lines to growth inhibition by both BRAF and MEK inhibitors, largely due to inhibition of apoptosis induction (104,112,117-119). The presence of a deletion or decreased copy number of the PTEN gene was associated with shorter PFS (median 4.5 vs. 8.0 months) in patients treated with dabrafenib, and lower PTEN protein expression was observed in non-responders than in responders to vemurafenib (76,104).
In addition to these genetic aberrations, the PI3K-AKT pathway appears to be critical to resistance mediated by RTKs to RAS-RAF-MEK-ERK pathway inhibitors (37,117). Notably, the activation of these RTKs in tumors and cell lines with acquired resistance is not mediated by mutations or amplifications of the genes that encode them, and therefore likely reflects epigenetic resistance mechanisms. Thus, there is a strong rationale to determine the clinical benefit of targeting this pathway in melanoma. There are multiple groups of inhibitors that target various effectors in the pathway (120-125). Preclinical studies have demonstrated that different genetic alterations in the pathway may correlate to sensitivity to specific classes of agents. For example, loss of PTEN has been associated with sensitivity to AKT inhibitors and isoform-specific PI3K inhibitors (70).
Additional molecular candidates from exome sequencing studies
The first complete genome sequencing of a human melanoma was reported in 2009 (126). This initial study identified 32,325 somatic base substitutions in a single patient, the majority of which likely resulted from the DNA-damaging effects of ultraviolet radiation. This daunting finding highlighted the critical need for exome sequencing studies of large numbers of melanomas in order to start to recognize recurrent events that are most likely to be meaningful. While the results of the melanoma TCGA effort are expected to be reported in 2014, initial whole exome studies have identify potentially significant events. A recurrent hotspot mutation in RAC1 was independently identified by two different groups by whole exome sequencing (8,9). The mutation was detected in 4-9% of melanomas. Expression of the resulting protein (RAC1P29S) demonstrated that the mutation increased MAPK activation, cell migration, and cell proliferation (8). New, statistically significant mutations in the coding regions of PPP6C, SNX31, TACC1, ARID2 and STK19 were also identified but their functional significance has not been reported (9).
TERT encodes the enzyme telomerase, which promotes cell survival by preventing DNA loss at the end of chromosome during cell division. Whole exome sequencing analysis did not detect any significant mutations in the coding region of the TERT gene. However, two recurrent hotspot mutations (C228T and C250T) were identified in the promoter region upstream of the gene (127-129). These mutations did not affect the sequence of the TERT protein, but they created a new binding site for transcription factors that can lead to increased TERT expression. These two mutations were mutually exclusive, and overall were detected in ~85% of cutaneous melanomas. The clinical associations and therapeutic significance of these mutations is currently unknown (Table 1).
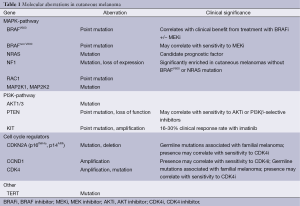
Full table
Molecular markers and resistance to BRAF inhibitors
The regulatory approval of vemurafenib [2011], dabrafenib [2013], and trametinib [2013] for metastatic melanoma patients with BRAFV600 mutations reflects the rapidly changing clinical landscape of melanoma. While these agents represent a clear advance compared to the only previously-approved cytotoxic agent (dacarbazine), their clinical benefit is limited markedly by resistance. As noted above, a number of pre-treatment factors have been associated with inferior outcomes with these agents, including: BRAFV600K mutations (versus BRAFV600E); loss of PTEN and CDKN2A; and amplification and/or gain of function mutations in CDK4 and CCND1. These associations support the rationale to develop combinatorial approaches that target these genes or their associated pathways (120,123,130,131).
In addition to pre-existing alterations that cause de novo resistance, a number of new alterations that are present at the time of disease progression have also been identified (Table 2). Notably, to date all progressing tumors and cell lines selected in vitro for resistance have demonstrated the continued presence of the same BRAFV600 mutation that was present prior to treatment (132). While no mutations in the BRAF gene have been identified, increased copy number of the BRAFV600 mutant allele has been identified in ~20% of patients at the time of progression on BRAF inhibitors (133,134). Testing of BRAF inhibitor (BRAFi)-resistant cell lines with this alteration demonstrated that growth inhibition could be achieved by simply using higher concentrations of BRAFi (135). Alternative splicing that causes expression of a smaller BRAFV600 protein has been detected in 15-20% of progressing patients (136). In contrast to the effects of BRAF amplification, the truncated BRAF proteins form dimers so efficiently that their effects cannot be overcome by increased dosing of BRAFi. However, this mechanism of resistance (MOR) does retain sensitivity to MEK or ERK inhibition. MEK and/or ERK inhibition are also predicted to overcome resistance mediated by the presence of a new activating mutation in NRAS or other RAS family members (132). In contrast to the mutual exclusivity observed in treatment-naïve patients, multiple studies have detected acquired NRAS hotspot mutations in 20-25% in progressing tumors after BRAFi treatment (132). Mutations in MEK1 and MEK2 that mediate resistance have also been identified in progressing lesions (134). Some of these mutations are predicted to also cause resistance to MEK inhibitors, but they appear to be sensitive to ERK inhibition.
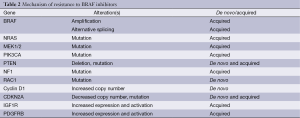
Full table
Overall, 50-70% of melanomas with acquired resistance to BRAF inhibitors have been found to harbor alterations that are predicted to re-activate signaling through the RAS-RAF-MEK-ERK pathway (133,134). Despite this, combined treatment with BRAF and MEK inhibitors has achieved clinical responses in only ~15% of patients who had previously progressed on single-agent BRAF inhibitor therapy (70,137). This lack of efficacy may be due in part to the presence of molecular events that cause concurrent activation of other signaling pathways. Both focused and whole exome sequencing studies have demonstrated that alterations predicted to activate the PI3K-AKT pathway are frequently detected in progressing lesions, often in the presence of concurrent acquired RAS-RAF-MEK-ERK alterations (133,134). In addition to genetic mutations (i.e., PTEN, PIK3CA, PIK3R1), increased expression of RTKs by epigenetic mechanisms has also been detected in these lesions (122,132). Preclinical studies support that combined inhibition of the PI3K pathway with MEK/ERK inhibitors may be required to achieve significant inhibition of tumor growth and survival in melanomas with such events. The detection of acquired mutations in CDKN2A and RAC1 also support the rationale for exploring inhibitors against those targets (133,134).
Another factor that may contribute to the refractoriness of BRAFi-resistant melanomas is the heterogeneity of these lesions (138,139). Isolated reports have demonstrated that different NRAS mutations may be present in different tumors from single patients, or may be present in only a portion of progressing lesions (76,132,140). A more recent study that performed whole exome sequencing on more than one progressing tumor in 16 patients found that 75% of the patients had evidence of different MORs in their different progressing tumors (141). Such findings suggest that broad and/or combinatorial treatment strategies may be required in the majority of BRAFi-resistant patients in order to overcome the multitude of resistance mechanisms that may be present.
Molecular testing and immunotherapy
High dose bolus interleukin-2 (HD IL-2) immunotherapy was approved for the treatment of patients with metastatic melanoma in 1998. This approval was not preceded by any randomized trials that demonstrated superior outcomes to other therapies. Instead, it was largely based upon the demonstration that ~5% of patients treated with HD IL-2 achieved durable (>5 year) disease control and survival, which no other therapies at that time had achieved (142,143). Over time, several breakthroughs in the understanding of the regulation of anti-tumor immune response have led to several new immunotherapy approaches for melanoma. Ipilimumab, an antibody that blocks the inhibitor CTLA-4 receptor on the surface of T-cells, was approved for the treatment of patients with metastatic melanoma in 2011 based on randomized trials that demonstrated significant improvements in PFS and OS (144,145). A recent pooled analysis of over 4,000 patients that had been enrolled on trials with ipilimumab found a 3-year OS rate of 21%, with a similar survival rate among patients with 5 and 10 year follow-up (146). More recently, antibodies against the inhibitory PD-1 receptor and its ligand PD-L1 have shown clinical response rates of 30-60% in early phase clinical trials (147-149). Other immunotherapies, such as adoptive cell transfer (ACT) of tumor infiltrating lymphocytes (TIL), have similarly demonstrated clinical responses in patients, many of which are very durable (>10 years) (150-152).
While these advances are impressive, the clinical use of immunotherapy for melanoma would be strengthened by the development of predictive biomarkers for these agents. There is growing evidence that mutational analysis may have utility in this area (153,154). Two different groups have reported that the presence of an activating NRAS mutation is associated with increased responsiveness to immunotherapies (155,156). Analysis of both preclinical specimens and patient biopsies have also demonstrated that BRAF inhibitor treatment increases the expression of melanocytic antigens on the surface of melanoma cells, resulting in improved recognition by T cells (157,158). A recent analysis has also demonstrated that loss of PTEN function results in the production of cytokines that dampen the antitumor response (159). Together these findings support the rationale to integrate analysis of immunological effects in targeted therapy trials, and the analysis of oncogenes in immunotherapy trials.
In addition to studies of functional mutations, it is also possible that somatic mutations may result in neoantigens that can be exploited for anti-tumor immune response. Initial studies of tumors from patients who have responded to TIL therapy have implicated immune recognition of antigens generated by somatic mutations in the clinical responses (160,161). Additional studies are warranted to improve our understanding of how often such mutations are relevant to responses to other immunotherapies, and perhaps to immune surveillance in early-stage melanoma patients.
Summary
The ability and need to genotype melanoma is now a clinical reality. At this time, the indication for molecular testing in cutaneous melanoma patients is clearest for tests for BRAFV600 mutations due to the availability of FDA-approved agents for patients with these mutations. However, as available data supports that most patients treated with single-agent BRAF inhibitor therapy will progress within one year, there is a strong rationale to expand the genotyping of these patients to identify and prioritize combinatorial clinical trials/approaches for patients. Similarly, the identification of multiple mechanisms of acquired resistance to BRAF inhibitors, and the specific therapeutic strategies that overcome each of them, suggests that molecular testing after disease progression may have clinical benefit. Testing for NRAS mutations in treatment-naïve patients adds confidence to negative test results for BRAFV600 mutations, and is also relevant as multiple new clinical trials open targeting this gene specifically. Impressive clinical responses have also been observed with FDA-approved agents in metastatic melanoma patients with BRAFNon-V600 (trametinib) and c-KIT (imatinib) mutations, although these agents have not gained specific regulatory approval for these indications to date. Finally, investigations are ongoing to determine if mutational testing will help to identify patients most likely to benefit from immunotherapies.
These findings demonstrate the marked advances that have been made in the understanding and treatment of melanoma in the last decade. However, a number of challenges and opportunities remain. Notably, while DNA-based testing has been clinically validated, at this time RNA- and protein-based molecular assays remain in developmental and/or testing phases. The recent studies of tumors that have progressed on BRAF inhibitor treatment highlight that effective therapies may affect both the nature and patterns of molecular events in this disease (i.e., co-occurrence of BRAFV600 and NRAS mutations). The identification of heterogeneous patterns of molecular changes between different tumors in individual patients, and even within individual tumors, will also need to be evaluated in the development of new markers. Despite these challenges, the progress that has been made already in a short period of time supports the enthusiasm for continued development of molecular testing approaches to further improve the management, survival, and quality of life for patients with this highly aggressive disease.
Acknowledgements
Funding: Michael A. Davies acknowledges funding support from the MD Anderson Goodfellow Scholar Endowment, NIH/NCI 1R01CA154710-01, Cancer Prevention Research Institute of Texas (CPRIT) R01, Melanoma Research Alliance Team Science Award.
Disclosure: Michael A. Davies has received research support from GlaxoSmithKline, Genentech, AstraZeneca, Oncothyreon, and Myriad. Michael A. Davies has served on advisory boards for GlaxoSmithKline, Genentech, and Novartis.
References
- Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214-8. [PubMed]
- Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 2012;485:502-6. [PubMed]
- Nathanson KL. Using genetics and genomics strategies to personalize therapy for cancer: focus on melanoma. Biochem Pharmacol 2010;80:755-61. [PubMed]
- Kudchadkar R, Gibney G, Sondak VK. Integrating molecular biomarkers into current clinical management in melanoma. Methods Mol Biol 2014;1102:27-42. [PubMed]
- Haq R, Fisher DE. Targeting melanoma by small molecules: challenges ahead. Pigment Cell Melanoma Res 2013;26:464-9. [PubMed]
- Normanno N, Rachiglio AM, Roma C, et al. Molecular diagnostics and personalized medicine in oncology: challenges and opportunities. J Cell Biochem 2013;114:514-24. [PubMed]
- De Luca A, Maiello MR, D’Alessio A, et al. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets 2012;16 Suppl 2:S17-27. [PubMed]
- Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet 2012;44:1006-14. [PubMed]
- Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251-63. [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [PubMed]
- Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum Mutat 2007;28:578-88. [PubMed]
- Jakob JA, Bassett RL, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012;118:4014-23. [PubMed]
- Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol 2011;164:776-84. [PubMed]
- Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 2011;29:1239-46. [PubMed]
- Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855-67. [PubMed]
- Garnett MJ, Rana S, Paterson H, et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell 2005;20:963-9. [PubMed]
- Tsao H, Goel V, Wu H, et al. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Investig Dermatol 2004;122:337-41. [PubMed]
- Goel VK, Lazar AJ, Warneke CL, et al. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol 2006;126:154-60. [PubMed]
- Grimaldi AM, Cassidy PB, Leachmann S, et al. Novel approaches in melanoma prevention and therapy. Cancer Treat Res 2014;159:443-55. [PubMed]
- Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst 2003;95:1878-90. [PubMed]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135-47. [PubMed]
- Handolias D, Salemi R, Murray W, et al. Mutations in KIT occur at low frequency in melanomas arising from anatomical sites associated with chronic and intermittent sun exposure. Pigment Cell Melanoma Res 2010;23:210-5. [PubMed]
- Greaves WO, Verma S, Patel KP, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn 2013;15:220-6. [PubMed]
- Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009;457:599-602. [PubMed]
- Wong CW, Fan YS, Chan TL, et al. BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J Clin Pathol 2005;58:640-4. [PubMed]
- Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19-20. [PubMed]
- Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J Natl Cancer Inst 2013;105:917-9. [PubMed]
- Omholt K, Platz A, Kanter L, et al. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res 2003;9:6483-8. [PubMed]
- Jakob JA, Bassett RL Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012;118:4014-23. [PubMed]
- Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol 2011;29:1239-46. [PubMed]
- Devitt B, Liu W, Salemi R, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res 2011;24:666-72. [PubMed]
- Meckbach D, Bauer J, Pflugfelder A, et al. Survival according to BRAF-V600 tumor mutations - an analysis of 437 patients with primary melanoma. PLoS One 2014;9:e86194. [PubMed]
- Menzies AM, Haydu LE, Visintin L, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res 2012;18:3242-9. [PubMed]
- Bucheit AD, Syklawer E, Jakob JA, et al. Clinical characteristics and outcomes with specific BRAF and NRAS mutations in patients with metastatic melanoma. Cancer 2013;119:3821-9. [PubMed]
- Dienstmann R, Tabernero J. BRAF as a target for cancer therapy. Anticancer Agents Med Chem 2011;11:285-95. [PubMed]
- Tap WD, Gong KW, Dering J, et al. Pharmacodynamic characterization of the efficacy signals due to selective BRAF inhibition with PLX4032 in malignant melanoma. Neoplasia 2010;12:637-49. [PubMed]
- Ribas A, Flaherty KT. BRAF targeted therapy changes the treatment paradigm in melanoma. Nat Rev Clin Oncol 2011;8:426-33. [PubMed]
- Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010;464:427-30. [PubMed]
- Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010;464:431-5. [PubMed]
- Halaban R, Zhang W, Bacchiocchi A, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res 2010;23:190-200. [PubMed]
- Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012;380:358-65. [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [PubMed]
- Falchook GS, Long GV, Kurzrock R, et al. Dabrafenib in patients with melanoma, untreated brain metastases, and other solid tumours: a phase 1 dose-escalation trial. Lancet 2012;379:1893-901. [PubMed]
- Ascierto PA, Minor D, Ribas A, et al. Phase II trial (BREAK-2) of the BRAF inhibitor dabrafenib (GSK2118436) in patients with metastatic melanoma. J Clin Oncol 2013;31:3205-11. [PubMed]
- Long GV, Trefzer U, Davies MA, et al. Dabrafenib in patients with Val600Glu or Val600Lys BRAF-mutant melanoma metastatic to the brain (BREAK-MB): a multicentre, open-label, phase 2 trial. Lancet Oncol 2012;13:1087-95. [PubMed]
- Klein O, Clements A, Menzies AM, et al. BRAF inhibitor activity in V600R metastatic melanoma. Eur J Cancer 2013;49:1073-9. [PubMed]
- Ponti G, Tomasi A, Pellacani G. Overwhelming response to Dabrafenib in a patient with double BRAF mutation (V600E; V600M) metastatic malignant melanoma. J Hematol Oncol 2012;5:60. [PubMed]
- Ponti G, Pellacani G, Tomasi A, et al. The somatic affairs of BRAF: tailored therapies for advanced malignant melanoma and orphan non-V600E (V600R-M) mutations. J Clin Pathol 2013;66:441-5. [PubMed]
- Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107-14. [PubMed]
- Flaherty KT, Infante JR, Daud A, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694-703. [PubMed]
- Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855-67. [PubMed]
- Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell 2004;6:313-9. [PubMed]
- Smalley KS, Xiao M, Villanueva J, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene 2009;28:85-94. [PubMed]
- Dahlman KB, Xia J, Hutchinson K, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2012;2:791-7. [PubMed]
- Falchook GS, Lewis KD, Infante JR, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. Lancet Oncol 2012;13:782-9. [PubMed]
- Sen B, Peng S, Tang X, et al. Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med 2012;4:136ra70.
- Palanisamy N, Ateeq B, Kalyana-Sundaram S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med 2010;16:793-8. [PubMed]
- Hutchinson KE, Lipson D, Stephens PJ, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res 2013;19:6696-702. [PubMed]
- Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res 2013;26:845-51. [PubMed]
- Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE 2004;2004:RE13.
- Colombino M, Capone M, Lissia A, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol 2012;30:2522-9. [PubMed]
- Yeh I, Bastian BC. Genome-wide associations studies for melanoma and nevi. Pigment Cell Melanoma Res 2009;22:527-8. [PubMed]
- Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol 2007;127:179-82. [PubMed]
- Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731-4. [PubMed]
- Ekedahl H, Cirenajwis H, Harbst K, et al. The clinical significance of BRAF and NRAS mutations in a clinic-based metastatic melanoma cohort. Br J Dermatol 2013;169:1049-55. [PubMed]
- Ellerhorst JA, Greene VR, Ekmekcioglu S, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res 2011;17:229-35. [PubMed]
- Heidorn SJ, Milagre C, Whittaker S, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell 2010;140:209-21. [PubMed]
- Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol 2013;14:249-56. [PubMed]
- Posch C, Moslehi H, Feeney L, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc Natl Acad Sci U S A 2013;110:4015-20. [PubMed]
- Kwong LN, Davies MA. Navigating the therapeutic complexity of PI3K pathway inhibition in melanoma. Clin Cancer Res 2013;19:5310-9. [PubMed]
- Kwong LN, Costello JC, Liu H, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med 2012;18:1503-10. [PubMed]
- Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res 2013;19:5320-8. [PubMed]
- Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2012;44:133-9. [PubMed]
- Emery CM, Vijayendran KG, Zipser MC, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A 2009;106:20411-6. [PubMed]
- Shi H, Moriceau G, Kong X, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer Discov 2012;2:414-24. [PubMed]
- Trunzer K, Pavlick AC, Schuchter L, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol 2013;31:1767-74. [PubMed]
- Bernards A, Settleman J. GAPs in growth factor signalling. Growth Factors 2005;23:143-9. [PubMed]
- Bikowska-Opalach B, Jackowska T. Neurofibromatosis type 1 - description of clinical features and molecular mechanism of the disease. Med Wieku Rozwoj 2013;17:334-40. [PubMed]
- Maertens O, Johnson B, Hollstein P, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov 2013;3:338-49. [PubMed]
- Whittaker SR, Theurillat JP, Van Allen E, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov 2013;3:350-62. [PubMed]
- Lennartsson J, Jelacic T, Linnekin D, et al. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells 2005;23:16-43. [PubMed]
- Huang S, Luca M, Gutman M, et al. Enforced c-KIT expression renders highly metastatic human melanoma cells susceptible to stem cell factor-induced apoptosis and inhibits their tumorigenic and metastatic potential. Oncogene 1996;13:2339-47. [PubMed]
- Woodman SE, Davies MA. Targeting KIT in melanoma: a paradigm of molecular medicine and targeted therapeutics. Biochem Pharmacol 2010;80:568-74. [PubMed]
- Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340-6. [PubMed]
- Kong Y, Si L, Zhu Y, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res 2011;17:1684-91. [PubMed]
- Abu-Abed S, Pennell N, Petrella T, et al. KIT gene mutations and patterns of protein expression in mucosal and acral melanoma. J Cutan Med Surg 2012;16:135-42. [PubMed]
- Papaspyrou G, Garbe C, Schadendorf D, et al. Mucosal melanomas of the head and neck: new aspects of the clinical outcome, molecular pathology, and treatment with c-kit inhibitors. Melanoma Res 2011;21:475-82. [PubMed]
- Abysheva SN, Iyevleva AG, Efimova NV, et al. KIT mutations in Russian patients with mucosal melanoma. Melanoma Res 2011;21:555-9. [PubMed]
- Zebary A, Omholt K, Vassilaki I, et al. KIT, NRAS, BRAF and PTEN mutations in a sample of Swedish patients with acral lentiginous melanoma. J Dermatol Sci 2013;72:284-9. [PubMed]
- Monsel G, Ortonne N, Bagot M, et al. c-Kit mutants require hypoxia-inducible factor 1alpha to transform melanocytes. Oncogene 2010;29:227-36. [PubMed]
- Roth KG, Zabor EC, Colgan MN, et al. Prognostic implication of KIT mutations in melanoma. J Clin Oncol 2013;31:abstr 9049.
- Carvajal RD, Antonescu CR, Wolchok JD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA 2011;305:2327-34. [PubMed]
- Hodi FS, Corless CL, Giobbie-Hurder A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol 2013;31:3182-90. [PubMed]
- Guo J, Si L, Kong Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol 2011;29:2904-9. [PubMed]
- Meyle KD, Guldberg P. Genetic risk factors for melanoma. Hum Genet 2009;126:499-510. [PubMed]
- FitzGerald MG, Harkin DP, Silva-Arrieta S, et al. Prevalence of germ-line mutations in p16, p19ARF, and CDK4 in familial melanoma: analysis of a clinic-based population. Proc Natl Acad Sci U S A 1996;93:8541-5. [PubMed]
- Becker TM, Rizos H, Kefford RF, et al. Functional impairment of melanoma-associated p16(INK4a) mutants in melanoma cells despite retention of cyclin-dependent kinase 4 binding. Clin Cancer Res 2001;7:3282-8. [PubMed]
- Zuo L, Weger J, Yang Q, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996;12:97-9. [PubMed]
- Walker GJ, Flores JF, Glendening JM, et al. Virtually 100% of melanoma cell lines harbor alterations at the DNA level within CDKN2A, CDKN2B, or one of their downstream targets. Genes Chromosomes Cancer 1998;22:157-63. [PubMed]
- Young RJ, Waldeck K, Martin C, et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res 2014;27:590-600. [PubMed]
- Jonsson A, Tuominen R, Grafstrom E, et al. High frequency of p16(INK4A) promoter methylation in NRAS-mutated cutaneous melanoma. J Invest Dermatol 2010;130:2809-17. [PubMed]
- Zebary A, Omholt K, van Doorn R, et al. Somatic BRAF and NRAS mutations in familial melanomas with known germline CDKN2A status: a GenoMEL study. J Invest Dermatol 2014;134:287-90. [PubMed]
- Sotillo R, Garcia JF, Ortega S, et al. Invasive melanoma in Cdk4-targeted mice. Proc Natl Acad Sci U S A 2001;98:13312-7. [PubMed]
- Nathanson KL, Martin AM, Wubbenhorst B, et al. Tumor genetic analyses of patients with metastatic melanoma treated with the BRAF inhibitor dabrafenib (GSK2118436). Clin Cancer Res 2013;19:4868-78. [PubMed]
- Davies MA. Regulation, role, and targeting of Akt in cancer. J Clin Oncol 2011;29:4715-7. [PubMed]
- Markman B, Tao JJ, Scaltriti M. PI3K pathway inhibitors: better not left alone. Curr Pharm Des 2013;19:895-906. [PubMed]
- Omholt K, Krockel D, Ringborg U, et al. Mutations of PIK3CA are rare in cutaneous melanoma. Melanoma Res 2006;16:197-200. [PubMed]
- Davies MA, Stemke-Hale K, Tellez C, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer 2008;99:1265-8. [PubMed]
- Davies MA. The role of the PI3K-AKT pathway in melanoma. Cancer J 2012;18:142-7. [PubMed]
- Aguissa-Touré AH, Li G. Genetic alterations of PTEN in human melanoma. Cell Mol Life Sci 2012;69:1475-91. [PubMed]
- Davies MA, Gershenwald JE. Targeted therapy for melanoma: a primer. Surg Oncol Clin N Am 2011;20:165-80. [PubMed]
- Xing F, Persaud Y, Pratilas CA, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene 2012;31:446-57. [PubMed]
- Haluska FG, Tsao H, Wu H, et al. Genetic alterations in signaling pathways in melanoma. Clin Cancer Res 2006;12:2301s-7s. [PubMed]
- Tsao H, Goel V, Wu H, et al. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol 2004;122:337-41. [PubMed]
- Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene 2003;22:3113-22. [PubMed]
- Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009;41:544-52. [PubMed]
- Paraiso KH, Smalley KS. Making sense of MEK1 mutations in intrinsic and acquired BRAF inhibitor resistance. Cancer Discov 2012;2:390-2. [PubMed]
- Deng W, Gopal YN, Scott A, et al. Role and therapeutic potential of PI3K-mTOR signaling in de novo resistance to BRAF inhibition. Pigment Cell Melanoma Res 2012;25:248-58. [PubMed]
- Gopal YN, Deng W, Woodman SE, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res 2010;70:8736-47. [PubMed]
- Atefi M, von Euw E, Attar N, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS One 2011;6:e28973. [PubMed]
- Shull AY, Latham-Schwark A, Ramasamy P, et al. Novel somatic mutations to PI3K pathway genes in metastatic melanoma. PLoS One 2012;7:e43369. [PubMed]
- Villanueva J, Vultur A, Lee JT, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 2010;18:683-95. [PubMed]
- Greger JG, Eastman SD, Zhang V, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther 2012;11:909-20. [PubMed]
- Kwong LN, Davies MA. Targeted therapy for melanoma: rational combinatorial approaches. Oncogene 2014;33:1-9. [PubMed]
- Davies MA, Samuels Y. Analysis of the genome to personalize therapy for melanoma. Oncogene 2010;29:5545-55. [PubMed]
- Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2010;463:191-6. [PubMed]
- Huang FW, Hodis E, Xu MJ, et al. Highly recurrent TERT promoter mutations in human melanoma. Science 2013;339:957-9. [PubMed]
- Horn S, Figl A, Rachakonda PS, et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013;339:959-61. [PubMed]
- Vinagre J, Almeida A, Populo H, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun 2013;4:2185. [PubMed]
- Carlino MS, Todd JR, Gowrishankar K, et al. Differential activity of MEK and ERK inhibitors in BRAF inhibitor resistant melanoma. Mol Oncol 2014. [Epub ahead of print]. [PubMed]
- Bucheit AD, Davies MA. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochem Pharmacol 2014;87:381-9. [PubMed]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973-7. [PubMed]
- Van Allen EM, Wagle N, Sucker A, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014;4:94-109. [PubMed]
- Wagle N, Van Allen EM, Treacy DJ, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 2014;4:61-8. [PubMed]
- Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun 2012;3:724. [PubMed]
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387-90. [PubMed]
- Sosman JA, Daud A, Weber JS, et al. BRAF inhibitor (BRAFi) dabrafenib in combination with the MEK1/2 inhibitor (MEKi) trametinib in BRAFi-naive and BRAFi-resistant patients (pts) with BRAF mutation-positive metastatic melanoma (MM). J Clin Oncol 2013;31:abstr 9005.
- Yancovitz M, Litterman A, Yoon J, et al. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in primary and metastatic melanoma. PLoS One 2012;7:e29336. [PubMed]
- Rizos H, Menzies AM, Pupo GM, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma; spectrum and clinical impact. Clin Cancer Res 2014;20:1965-77. [PubMed]
- Sensi M, Nicolini G, Petti C, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene 2006;25:3357-64. [PubMed]
- Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014;4:80-93. [PubMed]
- Atkins MB, Kunkel L, Sznol M, et al. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J Sci Am 2000;6 Suppl 1:S11-4. [PubMed]
- Atkins MB, Lotze MT, Dutcher JP, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol 1999;17:2105-16. [PubMed]
- Robert C, Thomas L, Bondarenko I, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011;364:2517-26. [PubMed]
- Hodi FS, O’Day SJ, McDermott DF, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711-23. [PubMed]
- Hodi FS. European Cancer Congress 2013 (ESMO-ECCO-ESTRO). Abstract LBA24. Presented September 28, 2013.
- Brahmer JR, Tykodi SS, Chow LQ, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455-65. [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [PubMed]
- Hamid O, Robert C, Daud A, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013;369:134-44. [PubMed]
- Rosenberg SA, Yang JC, Sherry RM, et al. Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 2011;17:4550-7. [PubMed]
- Hinrichs CS, Rosenberg SA. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 2014;257:56-71. [PubMed]
- Radvanyi LG, Bernatchez C, Zhang M, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res 2012;18:6758-70. [PubMed]
- Ott PA, Henry T, Baranda SJ, et al. Inhibition of both BRAF and MEK in BRAF(V600E) mutant melanoma restores compromised dendritic cell (DC) function while having differential direct effects on DC properties. Cancer Immunol Immunother 2013;62:811-22. [PubMed]
- Liu C, Peng W, Xu C, et al. BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin Cancer Res 2013;19:393-403. [PubMed]
- Joseph RW, Sullivan RJ, Harrell R, et al. Correlation of NRAS mutations with clinical response to high-dose IL-2 in patients with advanced melanoma. J Immunother 2012;35:66-72. [PubMed]
- Johnson DB, Lovly CM, Flavin M, et al. NRAS mutation: a potential biomarker of clinical response to immune-based therapies in metastatic melanoma (MM). J Clin Oncol 2013;31:abstr 9019.
- Boni A, Cogdill AP, Dang P, et al. Selective BRAFV600E inhibition enhances T-cell recognition of melanoma without affecting lymphocyte function. Cancer Res 2010;70:5213-9. [PubMed]
- Frederick DT, Piris A, Cogdill AP, et al. BRAF inhibition is associated with enhanced melanoma antigen expression and a more favorable tumor microenvironment in patients with metastatic melanoma. Clin Cancer Res 2013;19:1225-31. [PubMed]
- Dong Y, Richards JA, Gupta R, et al. PTEN functions as a melanoma tumor suppressor by promoting host immune response. Oncogene 2013. [Epub ahead of print]. [PubMed]
- Lu YC, Yao X, Li YF, et al. Mutated PPP1R3B is recognized by T cells used to treat a melanoma patient who experienced a durable complete tumor regression. J Immunol 2013;190:6034-42. [PubMed]
- Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 2013;19:747-52. [PubMed]


