
Feasibility and clinical utility of endoscopic ultrasound guided biopsy of pancreatic cancer for next-generation molecular profiling
Introduction
With the development of next generation sequencing (NGS) technology, there has been a dramatic increase in our knowledge and accelerated understanding of the molecular pathology of various cancer types, including pancreatic cancer (PC). PC remains the most lethal solid tumour type in humans, with little improvement in outcome over the last few decades. There are few effective therapies available for patients with advanced disease and as a result PC is predicted to become the 2nd leading cause of cancer death in the West by 2025 (1). A major challenge of translating recent genomic and pre-clinical discoveries in PC into clinical practice for precision medicine has been the implementation of real time molecular profiling of patients to inform clinical decision making. NGS has revolutionised our ability to classify patients into potential responsive subgroups through molecular profiling, however, the majority of these studies were performed using early stage, non-metastatic resected specimens (2-7). Even though NGS from core biopsies of metastatic lesions can be done with high success rates in PC (8,9), a large proportion of patients present with metastatic lesions that are not amenable to percutaneous biopsy due to anatomical location or size. This presents a significant challenge to obtain tissue for molecular phenotyping in all patients with PC, particularly in patients with localised disease. Thus, there is an urgent need to develop strategies to safely acquire sufficient quality tissue suitable for NGS for the entire spectrum of PC (resectable, locally advanced, metastatic) to enable precision medicine opportunities in real world clinical practice.
There has been significant progress in the use of endoscopic ultrasound (EUS) in the diagnosis and management of PC in the last decade (10-13). The development of next generation fine needle core biopsy needles has increased the quality and quantity of tissue samples that are obtained even from low epithelial content tumours which is a histopathological characteristic of PC (14,15). This provides an alternative to percutaneous biopsy for patients presenting with de novo PC irrespective of clinical stage.
Many studies describing the use of tumour biopsies, including EUS guided, for therapeutic stratification in PC have failed to describe the patient denominator included in the studies from the outset (8,13,15). In addition, utilising diagnostic samples for molecular analysis, has been associated with high failure rates in many cancer types including PC in previous studies (16-19). Therefore, the real world feasibility and clinical utility of EUS guided biopsies to enable NGS is not thoroughly investigated.
To address these challenges with the aim of facilitating real world personalised clinical trials in all clinical stages of PC, we developed a clinical patient pathway embedding translational research that allows NGS molecular phenotyping using diagnostic EUS guided biopsies in parallel and complementary to the standard diagnostic process.
Methods
Patients presenting with a pancreatic mass suspicious of PC between 2016–2018 were referred to a fast-track, early assessment EUS clinic in a single institution (Glasgow Royal Infirmary). Ethical approval was obtained for collecting additional research biopsies from patients undergoing EUS guided biopsies for investigation of possible PC (Ethical approval number: 17/WS/0085). Patient consent included extra biopsies of pancreatic lesions if deemed appropriate by performing clinician. Matching venous blood was taken for normal genome analysis and freshly frozen. Upon EUS evaluation, lesions suspicious of PC, IPMN or pancreatic neuroendocrine tumour (PNET) was sampled for diagnosis and genome sequencing. This increased the number of EUS needle passes from 2 or 3 to 4 or 5 per patient. Where appropriate, liver metastases (n=2) and lymph node metastases (n=1) were taken for comparative sequencing. Samples were either fresh frozen or stored in formalin prior to DNA/RNA extraction (Figure S1). Formalin fixed paraffin embedded (FFPE) diagnostic samples were fixed in a commercial methanol fixative in the endoscopy room (PreservCyt®). Fresh frozen samples underwent cryosection, whilst FFPE samples underwent standard tissue section and stained with haematoxylin and eosin (H&E). Histopathological assessment of diagnostic and fresh frozen H&E slides was performed by a Specialist Pancreatic Pathologist. If the diagnostic specimen was inconclusive, the fresh frozen research sample was utilised for diagnostic purposes. Samples identified for sequencing underwent histological cellularity assessment and areas containing tumour epithelium was identified and marked on the H&E slide. In the case of fresh frozen samples, all samples underwent macro-dissection to enrich for tumour epithelium prior to DNA and RNA extraction. Library preparation, DNA/RNA extraction and nucleic acid sequencing are fully described in the supplementary material.

Results
Clinical implications of utilising EUS biopsies for NGS
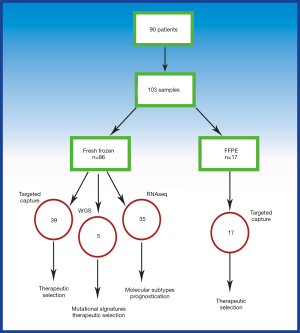
During the study period, 90 patients were used as a training set to develop the molecular profiling pathway to enable clinical feasibility (Table 1, Figure S2). The majority of patients (n=82, 91%) had a pathological diagnosis obtained from the initial EUS biopsy. The others were confirmed on repeat EUS (n=6) or laparoscopy (n=2) and 1 patient failed to obtain a histological diagnosis despite multiple attempts. The majority of patients were diagnosed with PC (n=65) and only 2 patients (2%) suffered morbidity during the admission of their diagnostic EUS. Both developed acute pancreatitis and subsequent temporary acute kidney injury after endoscopic retrograde cholangiopancreatography (ERCP).

Full table
EUS guided biopsies provide sufficient DNA and RNA yields for NGS
A major challenge to utilising EUS biopsies for molecular profiling is the perceived low DNA yields obtained from traditional fine needle aspirates. The DNA yield obtained from both fresh frozen and FFPE samples were sufficient using a variety of needles for targeted NGS, as well as whole genome sequencing (WGS) in the majority (73%) of patients with fresh frozen biopsy (Table 1). Fresh frozen biopsies allow sufficient RNA for whole transcriptome RNA sequencing in the majority of specimens (Table 1). Only 1 of the n=86 fresh frozen EUS guided biopsies provided insufficient DNA for sequencing, making the overall nucleic acid yield success rate of >98%.
To broaden the clinical utility and translational potential of this approach, a protocol was developed to test the diagnostic, FFPE samples to be utilised for NGS. A training set of 14 diagnostic FFPE samples were used for DNA extraction and all produced sufficient DNA (>100 ng) for targeted capture sequencing. Of these, 9 had matched fresh frozen biopsies that were processed and sequenced in parallel. In addition, FFPE diagnostic biopsy samples from 45 prospective patients enrolled in the PRECISION-Panc clinical trial platform demonstrated only 1 DNA extraction failure (Table 2). Of these, 27 were EUS biopsies and 19 percutaneous core biopsies of various metastatic lesions (Table 2). The only extraction failure was from a liver metastasis percutaneous biopsy due to insufficient material.

Full table
EUS guided biopsies can be utilised for targeted panel sequencing
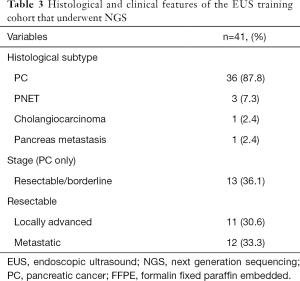
A cohort of consecutive patients (n=41) undergoing EUS biopsy underwent targeted capture, transcriptome (RNAseq) and whole genome (WGS) sequencing. The majority of patients had a diagnosis of PC (n=36, 87.8%) followed by pancreatic neuroendocrine tumour (n=3, 7.3%), Cholangiocarcinoma (n=1, 2.4%) and 1 patient with a pancreatic metastasis from a primary lung cancer (Table 3). Of the 36 patients with PC, 13 had borderline resectable/resectable disease (36.1%), 11 had locally advanced (30.6%) and 12 presented with metastatic (33.3%) disease (Table 3).
Full table
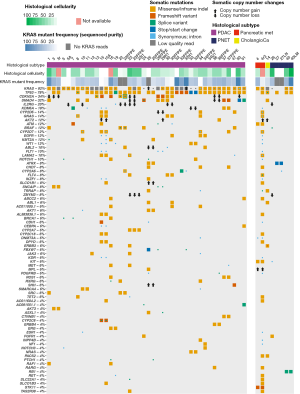
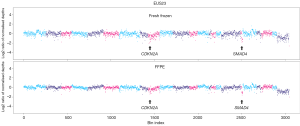
Targeted capture sequencing was performed following extraction of DNA (>50 ng). KRAS mutations were detected in 39 out of 42 samples (93%) from 25 out of 26 patients (96%). Only 1 sample was deemed a sequencing failure (EUS 16) based on quality control metrics but demonstrated a mutational profile consistent with PC (Figure 1). In 1 patient with no detectable KRAS mutation (EUS 22), there was a BRAF mutation suggesting that this was a true KRAS wild-type PC and not due to a sequencing failure (Figure 1). The mean allelic frequency of mutated KRAS appeared to correlate with histological tissue cellularity estimates, with very low KRAS frequency (<10%) being associated with histological cellularity <10% (Figure 1). Well known mutations in TP53 (78%), CDKN2A (34%) and SMAD4 (32%) were identified in keeping with previous studies (Figure 1) (2-4). Potentially actionable mutations were identified in a sub-group of patients, including ATM (12%) and BRCA1 (6%) (Figure 1). FFPE biopsies performed satisfactorily and provided sufficient DNA that is suitable for targeted capture sequencing (Table 2). Based on quality control analysis, these were above the thresholds required to call observed mutations (Table S1). Furthermore, the mutational profile obtained from FFPE samples were almost identical to the matched paired fresh frozen sample in both point mutation and copy number analyses (Figure 2).
Full table
EUS guided biopsies can be utilised for WGS
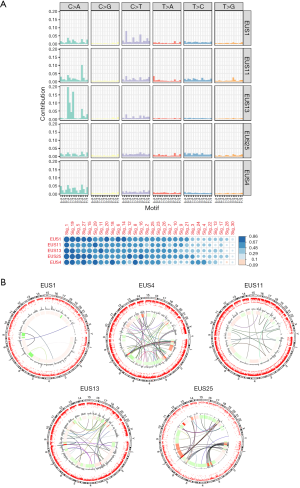
Due to the often-rapid emergence of resistance there is an urgent need to assess tumour evolution in response to therapeutics (20). In order to investigate these mechanisms including genomic rearrangements in addition to somatic mutations, WGS provides the most data. The feasibility of performing WGS using pre-treatment EUS biopsies was investigated with a proof of concept study of 5 selected samples based on molecular cellularity on panel sequencing (>25%), and available DNA quantity. In total, 31 of 43 patients (72%) had samples with sufficient quantity DNA for WGS, and 5 were selected as a proof of principle study with no sequencing failures. Mutational signature analysis revealed signatures previously described in PC including the COSMIC BRCA mutational signature (Figure 3). Circos plots allow visualisation of genomic re-arrangements and demonstrate the high number of structural variation events, as seen in EUS4 (Figure 3). These data suggest that fresh frozen EUS samples can be used for WGS to enable novel investigative techniques into the clonal evolution of PC.
RNA sequencing allows transcriptomic sub-typing of PC
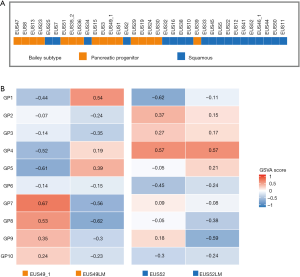
Molecular subtyping of tumours is becoming clinically relevant as therapeutic targets within subtypes are being identified and clinically tested (6). In order to allow treatment stratification based on molecular subtyping, the utility of RNAseq using EUS guided biopsies was investigated. Gene expression was normalised, and consensus clustering was performed on 35 PC based on gene programs described by Bailey et al. (2). We classified PC into the 2 main classes, squamous (n=19, 54%) and classical pancreatic (n=16, 46%) (6) (Figure 4). In the 2 patients with matching primary and liver metastases the primary and metastatic lesions cluster to the same subtype, despite some differences in gene programs (Figure 4). Albeit only in small numbers, this demonstrates that transcriptomic profiling of both primary and metastatic lesion is feasible using this approach and should be tested in further studies.
Discussion
Here we demonstrate that EUS guided biopsies can be utilised with high success rates in a routine clinical patient pathway for NGS analysis with the potential to enhance precision medicine and translational studies in PC. Importantly, acquiring extra biopsies at the same diagnostic EUS setting is associated with a high diagnostic rate without a significant increase in morbidity. At the same time, this eliminates the requirement for in-room cytopathology support at EUS. These results suggest that integrating research activities into routine clinical pathways is not detrimental to the quality of patient care and may further enhance it.
Panel sequencing from diagnostic EUS FNAs was previously shown to be feasible and able to identify potential actionable mutations (13). However, failure rates in using diagnostic samples was reported to be high and may result in a large proportion of patients requiring repeat research dedicated biopsies and subsequently being ineligible for personalised clinical trials. Our results demonstrate if sample acquisition is performed using a protocol tailored towards both diagnosis and NGS purposes, targeted capture sequencing can be performed with excellent success rates. Both fresh frozen and FFPE tissue provide sufficient DNA yields in almost all patients with high sequencing success rates (>90%). Using a number of paired fresh frozen and FFPE samples, our results demonstrate high concordance between both tissue acquisition and processing strategies. As a result, for ease of use and greater applicability across clinical units performing EUS, FFPE embedded biopsy tissue will be utilised for the PRECISION-Panc consortium (a clinical therapeutic development platform for pancreatic cancer, http://www.precisionpanc.org).
The ability to perform a range of NGS profiling and analysis in different preservative conditions will greatly improve the armamentarium available to study and treat PC. Developing a PC specific targeted capture based molecular assay that identifies signatures of therapeutic vulnerabilities such as DNA damage repair (DDR) and mismatch deficiency is a priority. WGS is significantly more expensive and requires much larger storage capacity, and as of yet has not demonstrated superiority for therapeutic selection. On the other hand, in well-designed studies focussed on clonal evolution of PC, WGS of EUS biopsies and subsequent resection specimen has potential to facilitate unique insights into the development of therapeutic resistance following neoadjuvant treatment strategies (Table S1). Furthermore, RNAseq of EUS guided biopsies can provide opportunities for investigating molecular subtypes in advanced disease and potentially direct therapy in future clinical trials.
The philosophy of PRECISION-Panc aims to provide clinical trial options for all patients with PC. The protocol developed here has been used successfully in >100 patients within the PRECISION-Panc master protocol. Ongoing evolution of this protocol and strategy is crucial as biomarkers of therapeutic response and molecular assays develop, to enable optimal patient selection for precision oncology in PC.
Supplementary
Endoscopic ultrasound biopsies: collection and extraction
Patients were sedated and received analgesia as standard (Midazolam, Fentanyl and local anaesthetic throat spray) for EUS and underwent initial endoscopy and ultrasound assessment of the pancreaticobiliary tract. Patients with evidence of a mass suspicious of a pancreaticobiliary neoplasm was biopsied as standard. This was followed by 1–3 additional samples for research purposes. Biopsies were taken using a fanning technique with a variety of EUS needles (discussed in Chapter 7). Diagnostic specimens were processed as standard using local collection protocols. This involved expelling all biopsies from the same lesion in a single pot of methanol based buffered preservative solution (ThinPrep Preservcyte, Hologic, inc. Cat No.: 85093-001). Additional research biopsies that were preserved in methanol fixative and embedded in FFPE was processed in a similar fashion. An additional venous blood sample (4–5 mLs) were collected in standard EDTA blood tubes (e.g., BD Vacutainer® K2EDTA tube, Cat no. KFK171) as a source of germline DNA.
EUS biopsies underwent cryopreservation to enable next generation sequencing including RNA and whole genome sequencing. This is a novel protocol designed by the PhD candidate and not previously described. Additional biopsies (1–3) were expelled in 5–10 mL of PBS in a 50 mL collection tube. This was gently swirled to allow excess blood to separate from the biopsy material. This was passed via a 70 μm nylon mesh cell strainer (Fisherbrand®, Cat No. 22363548) and the biopsy tissue transferred onto a metal histology mounting slide. This allowed the biopsies to all lie in a flat level plane, which enable cryosection at a later stage. The metal slide was transferred onto dry ice and the biopsies mounted in optimal cutting temperature (OCT) compound (VWR chemicals™, Cat No. 361603E) immediately. After the OCT has set, the mounted block and biopsies were removed from the mounting slide, placed in pre-labelled plastic cassette and transported to secure cold storage at −70 °C.
Fresh frozen EUS biopsies underwent histological analysis prior to DNA extraction, provided the diagnostic specimen was conclusive. In cases where uncertainty remained regarding the diagnosis, the fresh frozen specimens were reprocessed and embedded in FFPE to be used as diagnostic samples. Cryosections were performed by the Beatson Institute of Cancer research histopathology unit. Sections were stained with haematoxylin and eosin (H&E) followed by formal assessment by a Consultant Pathologist with an interest in pancreatic cancer. Regions with tumour epithelium were marked on H&E slides and histological cellularity determined. Macro dissection was performed to enrich for tumour epithelium in the frozen specimens. This involved overlaying the marked H&E slide with the OCT frozen block, whilst keeping the frozen tissue on dry ice. The corresponding marked areas were dissected using a fresh scalpel blade for DNA and RNA extraction.
DNA and RNA extractions were performed using the AllPrep® DNA/RNA micro Kit from Qiagen© (Cat. No. 80284). Briefly, on ice, 600 μL of RLT Plus solution (AllPrep® Micro Kit) was added to macro dissected tissue and disrupted using a rotor-stator homogenizer (Polytron® PT1200E, KINEMETICA) in a glass test tube. The lysate underwent freezing and thawing to allow complete lysis followed by centrifuging to separate the supernatant from tissue fragments. The supernatant was added to an AllPrep® DNA spin column, centrifuged and stored at 4 °C for extraction. Six hundred μL of Ethanol was added to the flow-through (containing RNA and protein) and added to an RNA spin column and centrifuged. This was followed by buffer washing of the spin column multiple times. RNA was isolated by eluting the RNA from the column using RNase-free water directly to the spin column (30–50 μL) and centrifuging for 1 minute at 8,000× g. DNA was isolated by buffer washing the DNA column and eluting the column with warmed elution buffer EB (AllPrep® Micro Kit). DNA and RNA were quantified using the Nanodrop® 2000 spectrophotometer. DNA and RNA were stored at −80 °C until sequencing.
Patients enrolled in the PRECISION-Panc master protocol that underwent molecular profiling from EUS biopsies had samples preserved in methanol fixative and embedded in FFPE. The commercial fixative used may vary from site to site, provided it is a methanol fixative (similar to ThinPrep PreservCyte, Hologic, inc. Cat No.: 85093-001). The PRECISION-Panc protocol requests patients to have a minimum of 3, but ideally 5, EUS biopsies collected and fixed in the same pot. Samples are then transferred to local pathology laboratory, where it is processed and embedded in formalin fixed paraffin embedded (FFPE) block for histological diagnosis followed by DNA and RNA extraction.
EUS biopsies are processed into FFPE blocks by retrieving all ‘micro-biopsies’ from the preservative pot using dedicated filter paper (CellPath™ tissuewrap). To avoid contamination, human fibrin or serum are not be used to make cell clots. These are next fixed for 12 to 24 hours in formalin and embedded as a paraffin block using standard histological techniques. Diagnostic H&E slide is taken, followed by cellularity estimation by dedicated Consultant Pathologist. An assessment on suitability for extraction and sequencing (sufficient tissue volume and tumour cellularity) is made by a consultant pathologist with significant experience in these techniques.
Extraction of FFPE biopsies
Formalin fixed EUS biopsies underwent DNA extraction by the NHS Greater Glasgow & Clyde Molecular Genetics Laboratory. Extraction in a clinically approved facility was selected as this ensures appropriate quality control for clinical trial enrolment and future treatment stratification. Sample extraction is performed using 2–4 10 μM tissue curls using the Maxwell® 16 FFPE Plus LEV DNA Purification Kit (Cat No. AS1135). The Maxwell® 16 System offers automation and walk-away purification that saves time and labour by eliminating reagent preparation, pipetting and centrifugation steps. Briefly, samples are prepared by centrifuging tissue curls and adding Proteinase K and Incubation buffer (included in Maxwell® Kit Cat No. AS1135). This is incubated at 70 °C overnight followed by the addition of lysis buffer. The sample is now ready for DNA purification and is added to the Maxwell® FFPE Plus LEV DNA cartridge. Automated extraction and elution are performed using elution buffer supplied in the extraction kit.
Extraction of germline DNA from blood
Germline DNA was obtained by venous blood preserved in standard diagnostic EDTA blood tubes. DNA was extracted using the DNeasy Blood Mini kit from Qiagen© (Cat No. 69504). Briefly, 200 μL of whole blood is added to 20 μL of Qiagen protease in a 1.5 mL microcentrifuge and incubated at 56 °C for 10 minutes. Two hundred μL of Ethanol is added, and the mixture applied to a DNA spin column. This was centrifuged, followed by buffered washing of the DNA column. DNA was isolated by elution with buffer AE (DNeasy Blood Mini Kit, Qiagen©). DNA yield was quantified using the Nanodrop® 2000 spectrophotometer and stored at −80 °C until sequencing.
Library preparation and sequencing
Sequencing libraries were created with Lisa Evers (Laboratory Scientist) and the Glasgow Precision Oncology Laboratory sequencing team.
Whole-genome library preparation
Whole-genome libraries were generated using either the Illumina TruSeq DNA LT sample preparation kit (Illumina, Part No. FC-121–2001 and FC-121–2001) or the Illumina TruSeq DNA PCR-free LT sample preparation kit (Illumina, Part No. FC-121–3001 and FC-121–3002) according to the manufacturer’s protocols. If available, 1 µg of DNA was used as input for fragmentation to ~300 base pairs (bp). In the EUS sequencing cohort lower quantities of DNA (down to 500 ng) was used for whole genome sequencing. Quantification of libraries for clustering was performed using the KAPA Library Quantification Kit - Illumina/Universal (KAPA Biosystems, Part No. KK4824) in combination with the Life Technologies Viia 7 real time PCR instrument.
RNA sequencing library generation and sequencing
RNA sequencing libraries for patient derived cell lines were generated using TruSeq Stranded Total RNA kits (catalogue No. RS-122-2203). Due to the relative low input of the RNA obtained from EUS biopsy samples, RNA sequencing libraries for these were performed using the KAPA RNA HyperPrep kit with Riboerase (KAPABIOSYSTEMS©, KK8561) designed for small input samples on Illumina® systems. Depending on the sample size up to 1 µg of RNA was used to produce libraries. cDNA was synthesized from the enriched and fragmented RNA using Invitrogen’s SuperScript II Reverse Transcriptase (catalogue number 18064) and random primers. This was converted into double stranded DNA and subjected to 15 cycles of PCR to produce RNA-seq libraries ready for sequencing. Prior to sequencing, libraries were examined for quality and quantity using an Agilent BioAnalyser and Caliper’s LabChip GX (part No. 122000) instruments using the DNA High Sensitivity Reagent kit (product No. CLS760672).
Targeted, whole genome and RNA sequencing
Sequencing was performed by the Glasgow Precision Oncology Laboratory sequencing facility. This is a state-of-the-art purpose-built facility and has recently obtained Good Clinical Laboratory Practice (GCLP) accreditation to allow clinically valid sample sequencing. Sequencing was performed on Illumina platforms according to the manufacturer’s instructions. All sequencing runs were subjected to quality control according to approved Glasgow Precision Oncology Laboratory standard operating procedures.
Acknowledgements
We would like to thank the Glasgow Royal Infirmary Endoscopy department, and NHS Greater Glasgow and Clyde Biorepository for their assistance in this study.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Ethical approval was obtained for collecting additional research biopsies from patients undergoing EUS guided biopsies for investigation of possible PC (Ethical approval number: 17/WS/0085).
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47-52. [Crossref] [PubMed]
- Biankin AV, Waddell N, Kassahn KS, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491:399-405. [Crossref] [PubMed]
- Waddell N, Pajic M, Patch AM, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495-501. [Crossref] [PubMed]
- Moffitt RA, Marayati R, Flate EL, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168-78. [Crossref] [PubMed]
- Collisson EA, Bailey P, Chang DK, et al. Molecular subtypes of pancreatic cancer. Nat Rev Gastroenterol Hepatol 2019;16:207-20. [Crossref] [PubMed]
- Collisson EA, Sadanandam A, Olson P, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17:500-3. [Crossref] [PubMed]
- Aguirre AJ, Nowak JA, Camarda ND, et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov 2018;8:1096-111. [Crossref] [PubMed]
- Aung KL, Fischer SE, Denroche RE, et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res 2018;24:1344-54. [Crossref] [PubMed]
- Bang JY, Hebert-Magee S, Navaneethan U, et al. EUS-guided fine needle biopsy of pancreatic masses can yield true histology: results of a randomised trial. Gut 2018;67:2081-4. [Crossref] [PubMed]
- Catenacci DV, Chapman CG, Xu P, et al. Acquisition of Portal Venous Circulating Tumor Cells From Patients With Pancreaticobiliary Cancers by Endoscopic Ultrasound. Gastroenterology 2015;149:1794-803.e4. [Crossref] [PubMed]
- Chang DK, Nguyen NQ, Merrett ND, et al. Role of endoscopic ultrasound in pancreatic cancer. Expert Rev Gastroenterol Hepatol 2009;3:293-303. [Crossref] [PubMed]
- Gleeson FC, Kerr SE, Kipp BR, et al. Targeted next generation sequencing of endoscopic ultrasound acquired cytology from ampullary and pancreatic adenocarcinoma has the potential to aid patient stratification for optimal therapy selection. Oncotarget 2016;7:54526-36. [Crossref] [PubMed]
- Artifon EL, Guedes HG, Cheng S. Maximizing the Diagnostic Yield of Endoscopic Ultrasound-Guided Fine-Needle Aspiration Biopsy. Gastroenterology 2017;153:881-5. [Crossref] [PubMed]
- Valero V 3rd, Saunders TJ, He J, et al. Reliable Detection of Somatic Mutations in Fine Needle Aspirates of Pancreatic Cancer With Next-generation Sequencing: Implications for Surgical Management. Ann Surg 2016;263:153-61. [Crossref] [PubMed]
- Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non-small cell lung cancer. Cancer 2015;121:631-9. [Crossref] [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res 2016;22:5772-82. [Crossref] [PubMed]
- Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015;33:2753-62. [Crossref] [PubMed]
- Zill OA, Greene C, Sebisanovic D, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer Discov 2015;5:1040-8. [Crossref] [PubMed]
- Burrell RA, Swanton C. Tumour heterogeneity and the evolution of polyclonal drug resistance. Mol Oncol 2014;8:1095-111. [Crossref] [PubMed]


