
Sézary Syndrome, recent biomarkers and new drugs
Introduction
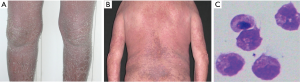
Sézary Syndrome (SS) is a rare aggressive leukemic variant of cutaneous T-cell lymphomas (CTCL), a spectrum of the malignant clonal proliferation of T lymphocytes with a predilection for skin involvement. CTCL presents commonly as mycosis fungoides (MF) (1,2). Most patients with SS are older adults, prevalent white males, with erythrodermia present in more than 80% of body surface area. Skin erythema that is often exfoliative, accompanied by severe pruritus, is present for months to years prior to diagnosis (Figure 1A,B) (4,5). Palmoplantar keratoderma, nail abnormalities, and alopecia are other commonly associated findings (5,6). Lymphadenopathy (≥1.5 cm in size) is common. Sezary cells (SS-cells) in blood, skin and lymph nodes, are the hallmark of SS (Figure 1C) and are usually CD4+ CD45RO+ (7). SS represents 3% of the CTCL with an increasing incidence rate (0.1–0.3/ 1,000,000) (1,8) and a medium age presentation of 60 years (9). Despite current treatments, the prognosis for patients is bad (10). Early diagnosis of SS is therefore key to achieve enhanced therapeutic responses (11). The diagnostic criteria are clonal T-cell receptor (TCR) rearrangement, peripheral Sezary cell counts of ≥1 G/L; CD4/CD8 ratio of ≥10; CD4+CD7− cells of ≥40%, or CD4+CD26− cells of ≥30%. The availability of anti-TCRVβ antibodies and of molecular techniques provides a suitable tool for SS diagnosis (12,13). Moreover, SS-cells express a phenotype of central memory T-cells (TCM) cells and are positive for cutaneous lymphocyte-associated antigen (CLA), and CC chemokine receptors (CCRs) (14). There are no specific biomarkers for neoplastic cells in SS patients and this represents a serious obstacle in evaluating early diagnosis and/or progression in this lymphoma (15). Biomarkers or biological markers, defined by Hulka and Wilcosky (16) as “cellular, biochemical or molecular alterations that are measurable in biological media such as human tissues, cells, or fluids”, include tools and technologies that can aid in understanding the diagnosis, progression, regression or outcome of clinical treatment of disease. Ideal diagnostic biomarkers are consistently and highly over-expressed in the malignant cells while absent or nearly absent in the normal cell population (17). While no single biomarker can be used to reliably diagnose SS, the combination of multiple markers has great potential and should be investigated prospectively.
In this review, we provide an overview of diagnostic, prognostic and therapeutic biomarkers in SS, discovered by high-throughput technologies over time. Finally, we discuss the most therapeutic agent available for the cure of this lymphoma and the ongoing interesting clinical trials.
Cell surface markers
The CLA and CCR4 expression are linked with the ability of SS-cells to localize to the skin, while the CCR7 expression is connected with SS entry into lymph nodes (7,18-20). TCR-Vβ detected either by molecularly or with specific monoclonal antibodies is so far a very good marker to assess the clonality of SS cells and also to follow tumor burden (13,21,22). Since as TCR-Vβ analysis is dependent on specific antibodies availability that is limited only to 70% of TCR-Vβ repertoire, the recent introduction of next-generation sequencing (NGS), for sequencing CD3/TCR, has improved the identification of malignant T cells (23,24). Furthermore, molecules such as the Natural Cytotoxicity Receptor (NCR) NKp46/NCR1 (25), CD85j/Ig-like transcript 2 (ILT2)-receptor (26), CD158k/KIR3DL2 receptor (27,28) and PD-1 (29) originally identified on natural killer (NK) cells; Ganglioside GD3(CD60), Integrin CD49d (30) and Syndecan 4 (SD-4) (31), present on activated normal T cells, were also found to be expressed at elevated levels in mainly high tumor burden SS patients. Cell-activation marker down/up-regulated in CD4 T cells from SS patients were also CD26, CD7 (3,20) and CD164, FCRL3 (32), CTLA-4 (33) Vimentin (34), respectively.
The intracellular protein T-Plastin, peculiarly expressed in malignant Sezary cells, remains so far usefulness as a diagnostic marker, because of its intracellular expression and the lacking of antibodies for flow cytometry (35,36). Gene expression does not always correlate with a presence on the cell surface because cell surface markers can be up- or down-regulated at the transcription or translation level.
NKp46/NCR1
The Natural Cytotoxicity Receptor NKp46 is an activating receptor principally expressed by NK cells. Bensussan et al. (25) showed that NKp46 is a cell surface marker, expressed in peripheral CD4+ lymphocytes of SS patients. The functional consequences of the engagement of NKp46 will be a severe inhibition of signals on proliferation CD3 dependent in Sezary cells. Their results revealed that NKp46, in combination with KIR3DL2/CD158k, could represent an available marker able to monitor the clinical course of this lymphoma (25).
CD85j/ILT2
CD85j/immunoglobulin (Ig)-like transcript 2 (ILT2) molecule is a member of the leukocyte immunoglobulin-like receptor (LIR) family (37). It transduces a negative signal able to inhibit stimulation of an immune response when it binds to MHC class I (MHC-I) molecules. Nikolova and colleagues (26) reported that CD85j is over-expressed Sezary cell lines and in circulating Sezary cells, with respect to CD4+ lymphocytes from healthy individuals. They also demonstrated that the triggering of CD85j leads to the recruitment of Src homology 2 domain-containing tyrosine phosphatase (SHP-1) and to the inhibition of proliferation induced by CD3/T-cell receptor stimulation in malignant CTCL cells. Finally, the authors demonstrated a lower susceptibility of Sezary cells expressing CD4+ILT2+ to anti-CD3 monoclonal antibody-induced cell death than the CD4+ILT2− lymphocytes.
KIR3DL2/CD158k
Another receptor found expressed in SS-cells is represented by KIR3DL2 who is part of the members of the killer-cell immunoglobulin-like receptors (KIRs). It was initially identified as expressed on NK cells and on rare circulating CD8+T cells (38). They have been named according to their two (KIR2D) or three (KIR3D) extracellular Ig-like domains. The KIRs have a long cytoplasmic tail (KIR-L) containing immune-receptor tyrosine-based inhibitory motifs or a short cytoplasmic tail (KIR-S) able to associates with molecules involved in activating signal (39). The KIR3DL2 expression is up-regulated upon activation of NK and T cells. In particular, KIR3DL2 ligation on activated T cells results in an anti-apoptotic effect and the production of IL-17 (40,41). KIR3DL2 has been identified as a cell surface marker in SS-cells in 2001 and its sensitivity for the diagnosis of Sézary Syndrome was confirmed in several studies (42-44). Indeed, the peripheral CD4+KIR3DL2+ T cells corresponded to the clonal malignant T-cell population identified by its unique TCR-Vβ rearrangement (45,46). Hurabielle et al. (47) demonstrated that KIR3DL2 expression is the most sensitive diagnostic SS-cells marker, and represents the best independent prognostic factor for the death of Sezary cells. Moreover, KIR3DL2 is relevant during the follow-up of this lymphoma (47). Recently, Roelens et al. (12) optimized a flow cytometry strategy demonstrating the heterogeneity of peripheral CD158k+ Sezary cells, being not exclusively of CD62L+ CCR7+ TCM phenotype. The authors characterized the phenotypic diversity of CD158k+T SS-cells in a large group of Sezary patients, including markers of T-cell subsets, like stem-cell and resident memory cells, elucidating a remarkable disparity between blood- and skin-derived CD158k+ T cells. Finally, skin-derived SS-cells show a more advanced maturation pattern with respect to circulating ones. The expression of CD69 and CD103 activation markers on the majority of skin-derived Sezary confirmed this data (12).
PD-1
Programmed death-1 (PD-1/CD279) belongs to the CD28/CTLA4 super-family and is expressed in a subset of activated T lymphocytes, monocytes dendritic cells, B-cells, and NK (29). The PD-1 exerts a key role in regulating immune response (48); it inhibits the TCR-mediated T-cell proliferation and cytokine secretion, by its ligands PD-L1, and PD-L2 (49). PD-1 is a part of a novel group of immune checkpoint able to promote apoptosis of antigen-specific T cells or to inhibit apoptosis in regulatory T cells (48) A new class of drugs, the PD-1 inhibitors, being developed for the treatment of several types of cancer, including CTCL (50).
Several studies reported the over-expression of PD-1/CD279 in various types of T-cell lymphomas, including SS (51-54). Samimi et al. (53) reported an increased expression of PD-1 in SS-cells than MF patients and healthy donor. A differential expression of PD-1 between SS and MF was observed in a study performed in skin biopsy by Cetinözman et al. (54); the authors provided a further support of the different entities of this two CTCL.
CD60 and CD49d
CD60 is a 9-O-acetylated form of the GD3 ganglioside expressed only by a part of T lymphocytes, the T memory effector cells (CD45RO+) (55). The ganglioside is an intracellular second lipid messenger able to induce apoptosis in lymphoid and myeloid cells. These effects are exerted by the way of FAS and FAS ligand, tumor necrosis factor (TNF-α) and/or amyloid signaling (56) and can be enriched through its de-O-acetylation. This mechanism could be useful for the development of new drugs therapies (30).
Another molecule involved in the migration of lymphocytes throughout the vessel wall is Integrin CD49d, also known as α4β1 or very late antigen-4 (VLA4): CD49d exerts this effect after it binds to vascular cell adhesion molecule-1 (VCAM-1), a cell adhesion molecule expressed on the activated endothelium. Scala et al. (30) evaluated the diagnostic and prognostic relevance of CD60 and CD49d expression on circulating CD4+T cells in 62 patients with SS. Over-expression of CD60 and down-regulation of CD49d predict the survival rate in these patients.
Syndecan-4
Syndecan-4 (SD-4) is a transmembrane heparan sulfate (HS) receptor expressed in a varies structure of HS in different cell types (57). Over-expression of specific epitopes of this receptor was found in Sezary cells (58). The constitutive over-expression of SD-4 in peripheral SS-cells distinguishes it from other leukemia or from other inflammatory cutaneous diseases (31).
CD26
CD26/dipeptidyl peptidase IV (DPPIV) is a proteolytic enzyme constitutively expressed in healthy peripheral T lymphocytes (59). This molecule is a cell activation marker up-regulated inside of mitogenic signals and down-regulated after the interplay with insulin-like growth factor II receptor (60). The expression of CD26 is losses in the cell surface of malignant T Sezary cells. CD26/DPPIV modulates the skin-homing chemotactic activity of many molecules, e.g., interferon-inducible T cell-α chemoattractant (I-TAC/CXCL11), a chemokine able to chemo-attractant T lymphocytes by CXCR3 (61). This mechanism could explain the ability of SS-cells CD26- for skin recruitment or for its intensification in peripheral blood (62). Narducci and colleagues confirmed the data on the abundance of skin-infiltrating Sezary T cells CD26−, analogous to peripheral Sezary cells (3). The authors demonstrated that the addition of soluble CD26 reduces the CXCR4-SDF-1 chemotaxis activity, abundantly signaling functionally active in skin homing of Sezary cells. Sokolowska-Wojdylo et al. reported that the CD26 is a very sensitive and specific marker, useful both in early detection and in therapeutic monitoring of SS disease (20).
CD164 and FCRL3
CD164/Sialomucin core protein 24, also known as endolyn functions as a cell adhesion molecule, is involved in the regulation of the adhesion, migration and proliferation, of hematopoietic progenitor cells (63). Sialomucin promotes myogenesis by enhancing CXCR4-dependent cell motility. FCRL3 or Fc Receptor-Like 3 is an immunoglobulin receptor which plays a role in the regulation of the immune system (64). Wysocka et al. demonstrated, for the first time, an over-expression of both FCRL3 and CD164 in Sezary T lymphocytes (32). The same authors demonstrated an inverse correlation between CD164 and CD26 expression and proposed the CD164 as a specific and sensitive marker for diagnosis, prognosis and staging of SS. Benoit B.M. and colleagues (15) demonstrated the high expression of CD164 in both peripheral and skin resident malignant CD4+ cells in CTCL patients. Additionally, the authors demonstrated an over-expression of KIR3DL2 and CD164 in 82% of Sezary patients.
CTLA-4/CD152
Cytotoxic T-lymphocyte antigen 4 (CTLA-4, CD152) is a T cell surface protein involved in immuno-regulation, inhibiting T cells activation or proliferation. A low expression of CTLA-4 is observed in un-stimulated cells whereas a higher expression is observed in stimulated cells (65-68). Gene fusion between CTLA-4 and CD28 molecule accompanied by over-expression was observed T Sezary cells (66,69,70). Gibson and colleagues (67) propose a mechanism via GATA3, a T-cell transcription factor, able to regulate Th2 differentiation, responsible for the over-expression of CTLA-4 in Sezary lymphocytes. The authors observed an up-regulation of CTLA-4 and GATA3 in normal T cell upon Bortezomib treatment, a proteasome inhibitor. The findings of CTLA4-CD28 gene fusion will be important for the therapeutic targeted strategy in SS (70).
Vimentin
Huet et al. (34) identified a monoclonal antibody termed SC5 that specifically recognizes vimentin (VIM), a type III intermediate cytoskeletal filament. It is expressed selectively by T lymphocytes isolated from healthy individuals upon activation, and by circulating Sézary Syndrome lymphocytes. Ortonne and colleagues (46) identified VIM at the surface membranes of Sezary cells and normally activated lymphocytes, conversely, in some patients the authors detected the presence of a nonmalignant circulating clone expressing high amounts of VIM and lacking CD158k.
Gene expression markers
The numerous expression studies conducted in SS identified a multitude of genes up- or down-regulated useful for differential diagnosis and progression in SS. Gene transcription studies have been performed extensively using blood samples or skin biopsies with quantitative reverse transcriptase PCR (qRT-PCR), microarray platform and transcriptome sequencing (RNA-Seq) (24,71). Nebozhyn et al. (72) developed a method, by qRT-PCR, to evaluate a set of five genes, concurrently: PLS3, STAT4 GATA3, TRAIL and CD1D. They demonstrated an efficiency of this set of genes of 90% in 49 SS patients and 65 healthy controls. Michel et al. (73) tested another panel of genes, KIR3DL2, NKp46 and PLS3, by qRT-PCR, obtaining an accuracy of 100% in 81 Sezary patients. Because KIR3DL2 and NKp46 were also expressed in NK cells of healthy controls, the authors proposed a flow cytometry analysis in combination with qRT-PCR, for this two markers. Litvinov et al. identified a set of 17 genes containing CCR4, IL2RA, TOX and STAT5A able to discriminate MF from SS patients (74). Boonk et al. (75) compared CD4+ T lymphocytes from 59 SS versus 19 erythrodermic inflammatory dermatosis patients analyzing copy number alterations data, gene expression and flow cytometry analysis. Malignant T SS-cells showed MYC gain (40%) and MNT loss (66%); up-regulation of EPHA4 (66%), DNM3 (75%), PLS3 (66%), and TWIST1 (69%); and down-regulation of STAT4 (91%). Loss of the CD26 (≥80% CD4+ T lymphocytes) and/or of the CD7 (≥40% CD4+ T lymphocytes) in combination with the aberrant expression of TWIST1, STAT4, PLS3 and DNM3 will be useful in the differential diagnosis between SS and patients affected by erythrodermic inflammatory dermatoses (100% specificity) (75). Recently, the same authors demonstrated, in SS patients, a direct correlation between over-expression of PLS3 and a favorable disease outcome; on the other hand, evaluated an inverse correlation between a higher leukocyte count and survival rate (76). Caprini et al. (77) were able to characterize 113 deregulated genes in lesioned chromosomal regions combining mapping and transcriptional data. The authors found important cancer-related genes such as members of the NF-kappaB pathway (TRAF2, BAG4, NKIRAS2, BTRC and PSMD3) that might explain its constitutive activation in CTCL (77). Moreover, they identified some genes like PIP5K1B and BUB3 that might exert critical roles in Sezary disease. Litvinov et al. (78) reviewed over 400 different studies to select 284 genes that were reported to play an important role in CTCL pathogenesis and progression. Their results underscore significant molecular heterogeneity with respect to gene expression between different patients and even within the same patients over time. Authors demonstrated that CCL18, CCL26, CCR4, FYB, T3JAM, MMP12, LEF1, LCK, ITK, GNLY, IL2RA, IL-26, IL-22, IL1F, BLK, CDO1, GTSF1, EED, THAP11, SYCP1, cTAGE1, POU5F1, POU2AF, STAT4, STAT5 and TOX could jointly be used as diagnostic and poor prognostic markers in CTCL patients, while SERPINB13, PSORS1C2, WIF1 and BCL7A were preferentially up-regulated in benign skin conditions and/or in indolent CTCL.
High-throughput technologies such as RNA sequencing, are an attractive approach for clinical diagnostic or staging of this lymphoma, allowing deeper characterization of targeted cancer genes that are frequently altered, at an affordable cost and in an efficient time frame.
Lefrançois et al. (79) compared RNA-Seq gene expression data obtained from FFPE samples collected earlier of 2008 with respect to those collected after 2009. Both analyses produced nearly identical trends and findings. Furthermore, the authors characterized a significant novel set of differentially expressed genes up-regulated in CTCL such as SMAD1, BIRC5, CASP1, BCL11A, IRF1, MAX and SELL and down-regulated such as THBS4, SERPINB3 and MDM4.
Iżykowska et al. (80) focused on those changes that affect the expression of the same gene in more than a patient and identified two genes, NCOR1 and CTBP1, which although the genomic alterations were over-expressed at mRNA level, in more SS patients. These genes are both involved in histone deacetylases (HDAC) complexes, able to regulate the HDAC activity. The destabilization of this complex alters gene expression in numerous human diseases (81), including SS. These results deserve attention because of the approved treatment of HDAC inhibitors in SS since 2006 (82).
MicroRNA and other regulatory RNA
MicroRNAs (miRNAs) are short non-coding RNA molecules involved in critical biological processes, such as proliferation, apoptosis, immune function, development, and the stress response through negative regulation of the stability and translation of target messenger RNAs (83,84). miRNAs are tissue/specific biomarkers with clinical relevance for tumor diagnosis and prognosis (84). MiRNAs microarray studies, conducted in SS identified, an aberrant expression of a significant number of miRNAs (85). Of note, several miRNA genes (>50%) are mapped in cancer-related regions or in fragile sites in SS patients (86), as for example miR21, miR486 and miR31 (87). Some of these miRNAs, which are over-expressed in SS, negatively modulate PTEN expression directly, such as miR214,miR21, miR106b and miR486, whereas others modulate PTEN expression indirectly, such as mi199a (88,89). Furthermore, Benoit et al. (15) evaluated as malignancy marker, in CD164+and CD164− SS cells, the expression level of FCRL3, TOX, miR21 and miR214. Worthy of note is the role played by the over-expression of miRNA155 in CTCL. Several studies elucidated its contribution to promoting inflammation and cancer development by way of the STAT signaling (83,90). On the other hand, several down-regulated miRNAs, such as miR342, miR31 and miR7 family members, may inhibit cancers by regulating genes involved in apoptosis, such as CDKN2A-B and TNFSF11 (85). Ballabio and colleagues (85) described also an increased level of miR223 in SS-cells with 91% specificity and 90% sensitivity compared with healthy controls. Ralfkiaer et al. (83) distinguished CTCL from benign skin diseases with a miRNA panel (miR155, miR203,miR205) with an accuracy of 95%.
Dusílková et al. (91) studied a system for detecting miRNAs in the plasma of patients affected by CTCL. MiRNAs levels measured in serum or plasma were very stable and resistant to RNA degradation and their detection is becoming promising for clinical monitoring. Based on the miR155 up-regulation and miR203/miR205 down-regulation (with 100% specificity and 94% sensitivity) their data provide evidence of a very efficient method able to evaluate the CTCL progression and to distinguish it from benign lesions.
Finally, Lee et al. (92) identified 12 long non-coding RNAs in three SS patient with miRNA-Seq demonstrating the presence of long non-coding RNAs in this lymphoma (Figure 2, microRNAs).
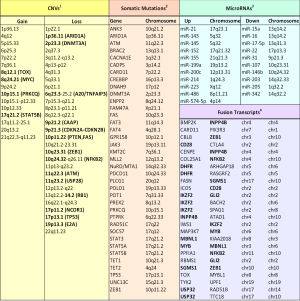
Copy number variation and somatic mutation markers
The genetic alterations in CTCL, in particular in SS, have been studied using cytogenetic and array-based methods for a long time. These studies identified extensive genetic instability with complex karyotypes but no highly recurrent translocations (95,96). A series of genomic studies identified large chromosomal regions affected by recurrent copy number alterations (CNAs). SS patients display significant CNAs (gains) in regions of already known and described oncogenes, like MYC (8q24.21), TOX (8q12.1) (95,97) and deletion in regions of suppressor genes like TP53 (17p13.1) (95,98), PTEN, FAS (10p11.22), RB1 (12p13.1), ZEB1 (10q23.31) (88,93,97,99), CDKN2A-CDKN2B (9p21.3) (100), E2A (19p13.3) DNMT3A (2p23.3) (80,101), USP28 (11q23.2), CAAP (9p21.2), NCOR1 (17p11.2) (80) and A20 (6q23.3) (102). In addition to the genes already reported, Vermeer and colleagues (95), observed losses of RPA1, HIC1, TCF8/ZEB1, DUSP5 and gain of STAT3/STAT5 and IL-2 (receptor) genes. Caprini et al. (77) using single nucleotide polymorphism and comparative genomic hybridization array confirmed previous studies with recurrent losses and gain regions, whereas identified novel genomic lesions recurring in >30% of tumors: loss of 9q13-q21.33 and gain of 10p15.3-10p12.2. Noteworthy, the same authors, observed that more than three recurrent chromosomal alterations (gain or loss) had shown a significant correlation with SS prognosis (77) (Figure 2, CNVs).
NGS is particularly suitable for the discovery of different types of genetic alterations ranging from pathogenic single nucleotide substitutions to large chromosomal aberrations in cancer genomes. In recent years, genome-wide studies using NGS have also provided a more comprehensive picture of the genetic landscape of CTCL (Figure 2, somatic mutations).
The majority of somatic single nucleotide substitutions observed in SS were C>T transitions (40–75%) (94,103), which were much less common in other hematological cancers. This mutational signature is regarded as caused by ultraviolet (UV) light exposure when occurring at NpCpG sites (104) or aging when occurring at NpCpC sites. In SS, both aging (43%) and UV radiation (30%) contribute to C>T transitions (94,105). At present, it is unclear whether exposure to UV light plays a causative role in the onset of the disease (106). Woollard and colleagues (107) described the molecular heterogeneity of CTCL tumor cells identifying more than 800 somatic non-synonymous mutations, including indels, stop-gain/loss, splice variants, and recurrent gene variants. The authors identified a new candidate driver gene in CTCL like POT1, TP53 and DNMT3A and confirmed previous literature data of alterations of TP53 and DNMT3A. Novel somatic mutations were identified in PLCG1 and BRCA2 detected in 11% and in 14 of SS patients, respectively. Chromosomal aberrations of PRKCQ, a gene involved in activation of TCR and NF-κB signaling, occurred in 20% of SS patients (107). Prasad et al. (69) identified four genes with recurrent somatic mutations, including stop-gain mutation, missense mutations, damaging somatic mutations, in a cohort of 15 SS patients: TP53, ITPR1, DSC1, and PKHD1L1. Moreover, the authors reported single somatic mutations in several genes such as PLCG1, STAT5B, GLI3, CARD11, NAV3, RIPK2, IL6, RAG2 and ITPR2, some of which previously described in CTCL: PLCG1, STAT5B, GLI3, CARD11 and NAV3 (93,94,104,108-111). Other genes with somatic point mutations were IL6, RIPK2, RAG2 (69). More recently, all CTCL samples with publicly available sequencing data have been reanalyzed applying uniform methods and metrics to generate a more representative overview of point mutation frequencies in putative driver genes (112). Park et al. (112) identified 55 putative driver genes in a genomic study conducted in 220 CTCLs. The authors discovered novel mutations involved in chromatin remodelling, such as BCOR, KDM6A, SMARCB1 and TRRAP; in immune surveillance, like CD58 and RFXAP; in signaling of MAPK kinase, like MAP2K1 and NF1; in signaling of NF-κB, such as PRKCB and CSNK1A1; in PI-3-Kinase signaling, like PIK3R1and VAV1;in RHOA/cytoskeleton remodeling (ARHGEF3); in RNA-splicing (U2AF1); in TCR signaling, such as PTPRN2and RLTPR and in T cell differentiation (RARA). Point mutations either predicted or confirmed to be gain-of-function were reported in JAK1 (0.9% of cases), JAK3 (2.7%), STAT3 (0.9%), and STAT5B (3.6%) (112). Recurrent mutations were also reported in four genes not previously identified in cancer: CK1α, PTPRN2, RARA, and RLTPR. Finally, they validate two putative oncogene CSNK1A1 and RLTPR recently described as involved in the TCR signaling (112).
Fusion transcript
Fusion genes are hybrid genes composed by parts of two or more genes, which origin from aberrant chromosomal or transcription rearrangements (113). Fusion genes might be responsible for inactivation of either tumor suppressor genes or activation for oncogenes. They could act as driver genes of tumor initiation and progression. Fusion genes will be useful as diagnostic and prognostic markers and represent an important class of targetable events to improve patient outcomes.
A well described gene-fusion in SS was CTLA4/CD28 transcript (70,94,108). Expressed in normal T cells, the CD28 is a molecule involved in a cascade of activation signals including cell proliferation and transcription of CTLA4. CTLA4, in turns, inhibits proliferation by opposing the effects of CD28 (114). In the fusion gene, the inhibitory cytoplasmic tail of CTLA4 was replaced by the activating tail of CD28. This fusion could provide a novel aberrant stimulatory mechanism contributing to oncogenic proliferation and survival (114). Sekulic et al. reported one SS-patient trial with CTLA4-blocking antibody Ipilimumab produced dramatic initial responses for first 2 months, but the response was limited and the patient died 3 months after the last dose (70). Wang et al. identified 41 in-frame fusion transcripts, by RNA-seq (94), 29 of them were successfully validated. In addition to CARD11-PIK3R3 fusion, they observed CD28-CTLA4/ICOS and IKZF2-GLI2 fusions. Prasad and colleagues, performing RNA-Seq in 15 SS, identified putative driver gene mutations implicated in the progression of this lymphoma (69). In addition, 86 potential fusion transcripts were identified in 10 Sezary samples (69). Some of these fusion events were successfully validated, such as TYK2-UPF1, COL25A1-NFKB2, FASN-SGMS1, SGMS1-ZEB1, SPATA21-RASA2, PITRM1-HK1, and BCR-NDUFAF6. Iżykowska et al. (80) identified 9 new fusion events in frame (EHD1-CAPN12, TMEM66-BAIAP2, MBD4-PTPRC, PTPRC-CPN2, MYB-MBNL1, TFG-GPR128, MAP4K3-FIGLA, DCP1A-CCL27, MBNL1-KIAA2018) in nine SS patients; 5 fusion transcripts were described as ectopic expression of fragments genes not expressed in normal T lymphocytes (BAIAP2, CPN2, GPR128, CAPN12, FIGLA). Those transcripts, except for one (TFG-GPR128) (115), have never been reported in the literature in SS (Figure 2, fusion transcripts).
Current therapy in Sézary Syndrome
The treatment of SS is primarily determined by disease extent and the impact on quality of life, prognostic factors and patient age/comorbidities an important principle in SS treatment is the preference for treatments that preserve, rather than suppress, the patient’s immune system. SS is associated with dysfunction of cellular immunity resulting in impaired response to infections and tumors, therefore the use of immunosuppressing therapies may lead to overwhelming infection. As infection is a common cause of death, another principle in SS treatment is vigilant surveillance for microbial colonization and infection (116).
First-line therapies for SS include extracorporeal photopheresis (ECP); subcutaneous interferon-α (IFN-α); oral bexarotene; and low-dose oral, subcutaneous, or intramuscular methotrexate. Many combination regimens are possible: bexarotene and IFN-α; bexarotene and ECP; IFN-α and ECP; bexarotene, IFN-α, and ECP; IFN-α and low-dose MTX; and IFN-α, low-dose MTX, and ECP. Chlorambucil and prednisone are also recommended (117). Adjuvant systemic agents comprehend antihistamines, doxepin, and gabapentin (6). Emollients and topical corticosteroids may be a helpful presence of pruritus.
Systemic therapy may also be combined with skin-directed therapy: psoralen plus ultraviolet A (PUVA) with bexarotene, IFN-α, and ECP; low-dose MTX and topical nitrogen mustard; PUVA and bexarotene; and total skin electron beam therapy (TSEBT) may be combined with ECP, IFN-α, and bexarotene. Because it is a radiosensitizer, MTX is not administered at the same time as TSEBT. Topical corticosteroids may be used in combination with any systemic therapy (6,118,119).
Second-line therapies for SS include single-agent chemotherapy (liposomal doxorubicin, gemcitabine, low-dose pralatrexate, pentostatin, chlorambucil, etoposide, cyclophosphamide, temozolomide, and high-dose MTX), HDACi (oral vorinostat and intravenous romidepsin), multiagent chemotherapy [fludarabine and cyclophosphamide; cyclophosphamide, doxorubicine, vincristine, prednisone (CHOP)], or targeted immunotherapy including brentuximab (anti-CD30), alemtuzumab (anti-CD52), and mogalumizumab (anti-CCR4) (117,119).
Although many treatments are available, most responses are partial and not durable, therefore the course of therapy in SS is that of successive treatments either alone or in combination. Furthermore, the effect of treatment on overall survival is unknown (6). An exception may be allogeneic bone marrow transplantation (BMT), which has provided durable complete remissions in SS, however, it is associated with high morbidity and mortality and is currently only considered in young, relatively healthy patients with advanced disease (120).
Current clinical trials with new drugs
The newly identified biomarkers and especially the genetic analysis combined with the availability in the cancer therapeutics field of new targeted therapies has opened the road to new therapeutic possibilities in these patients Several new agents for the treatment of SS are in development, including those that target KIR3DL2 (28), CD3 (a pan-T-cell marker), CD25 (IL-2 receptor, the target of denileukin diftitox), PD-1 receptor (an immune checkpoint targeted by pembrolizumab), and PI-3 kinase(a signal transducer inhibited by duvelisib). Second-line systemic agents include aprepitant, mirtazapine, and selective serotonin reuptake inhibitors (6). Food and Drug Administration (FDA)-approved JAK inhibitors such as Tofacitinib and Ruxolitinib make the idea of targeting malignant T cells driven by hyperactive JAK kinases attractive. Pharmacological inhibition of complex NF-κB by Bortezomib decreased its DNA-binding ability and induced cell death of SS cells in vitro (106,121-123). Two systemic HDAC inhibitors (HDACis), romidepsin (class I-specific HDACi) and vorinostat (pan-HDACi), are FDA approved for the treatment of CTCL. Trials with (novel) HDACis as single agents or in combinations are now underway in diverse cancer types, including hematologic and solid tumors, but have not yet reached the clinical (124). More details of the current clinical trials are indicated in Figure 3.
Conclusions
High-throughput methods provide invaluable insights into the molecular mechanisms or progression of this disease. The rare cancers are moving toward personalized medicine; the large scale of clinical trials are expanding and the identification of somatic alterations are driving the treatment decision. News insight molecular biomarkers allowed a partial stratification of SS patients implementing accurate diagnostic methods. Individual therapies are still deficient and the progress of the disease, with the manifestation of resistant clones, is very frequent and gradual. Patients with an advanced stage of disease should be inserted in multicenter clinical trials. Therapeutic strategies should maintain a quality of life that should be considered alongside response rates in clinical research. The characterization of new targetable biomarkers and the development of monoclonal antibodies seems to be increasingly promising for the cure of this lymphoma.
Acknowledgements
This study was partially supported by Associazione Italiana Ricerca sul Cancro (AIRC Grant to G Russo and MG Narducci); Fondi Ricerca Corrente from Italian Ministry of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood 2005;105:3768-85. [Crossref] [PubMed]
- Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch Dermatol 2007;143:854-9. [Crossref] [PubMed]
- Narducci MG, Scala E, Bresin A, et al. Skin homing of Sézary cells involves SDF-1-CXCR4 signaling and down-regulation of CD26/dipeptidylpeptidase IV. Blood 2006;107:1108-15. [Crossref] [PubMed]
- Wieselthier JS, Koh HK. Sézary syndrome: diagnosis, prognosis, and critical review of treatment options. J Am Acad Dermatol 1990;22:381-401. [Crossref] [PubMed]
- Kubica AW, Davis MDP, Weaver AL, et al. Sézary syndrome: a study of 176 patients at Mayo Clinic. J Am Acad Dermatol 2012;67:1189-99. [Crossref] [PubMed]
- Spicknall KE. Sézary syndrome-clinical and histopathologic features, differential diagnosis, and treatment. Semin Cutan Med Surg 2018;37:18-23. [Crossref] [PubMed]
- Campbell JJ, Clark RA, Watanabe R, et al. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood 2010;116:767-71. [Crossref] [PubMed]
- Bradford PT, Devesa SS, Anderson WF, et al. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood 2009;113:5064-73. [Crossref] [PubMed]
- Wilson LD, Hinds GA, Yu JB. Age, race, sex, stage, and incidence of cutaneous lymphoma. Clin Lymphoma Myeloma Leuk 2012;12:291-6. [Crossref] [PubMed]
- Li JY, Horwitz S, Moskowitz A, et al. Management of cutaneous T cell lymphoma: new and emerging targets and treatment options. Cancer Manag Res 2012;4:75-89. [PubMed]
- Kim EJ, Hess S, Richardson SK, Newton S, Showe LC, Benoit BM, et al. Immunopathogenesis and therapy of cutaneous T cell lymphoma. J Clin Invest 2005;115:798-812. [Crossref] [PubMed]
- Roelens M, Delord M, Ram-Wolff C, et al. Circulating and skin-derived Sézary cells: clonal but with phenotypic plasticity. Blood 2017;130:1468-71. [Crossref] [PubMed]
- Scala E, Narducci MG, Amerio P, et al. T Cell Receptor-Vβ Analysis Identifies a Dominant CD60+ CD26- CD49d- T Cell Clone in the Peripheral Blood of Sézary Syndrome Patients. J Invest Dermatol 2002;119:193-6. [Crossref] [PubMed]
- Berger CL, Edelson RL. Current Concepts of the Immunobiology and Immunotherapy of Cutaneous T Cell Lymphoma: Insights Gained through Cross-talk between the Clinic and the Bench. Leuk Lymphoma 2003;44:1697-703. [Crossref] [PubMed]
- Benoit BM, Jariwala N, O’Connor G, et al. CD164 identifies CD4+ T cells highly expressing genes associated with malignancy in Sézary syndrome: the Sézary signature genes, FCRL3, Tox, and miR-214. Arch Dermatol Res 2017;309:11-9. [Crossref] [PubMed]
- Hulka BS, Wilcosky T. Biological Markers in Epidemiologic Research. Arch Environ Health 1988;43:83-9. [Crossref] [PubMed]
- Kohnken R, Fabbro S, Hastings J, et al. Sézary Syndrome: Clinical and Biological Aspects. Curr Hematol Malig Rep 2016;11:468-79. [Crossref] [PubMed]
- Ferenczi K, Fuhlbrigge RC, Kupper TS, et al. Increased CCR4 Expression in Cutaneous T Cell Lymphoma. J Invest Dermatol 2002;119:1405-10. [Crossref] [PubMed]
- Sokolowska-Wojdylo M, Wenzel J, Gaffal E, et al. Circulating clonal CLA+ and CD4+ T cells in Sezary syndrome express the skin-homing chemokine receptors CCR4 and CCR10 as well as the lymph node-homing chemokine receptor CCR7. Br J Dermatol 2005;152:258-64. [Crossref] [PubMed]
- Sokolowska-Wojdylo M, Wenzel J, Gaffal E, et al. Absence of CD26 expression on skin-homing CLA+ CD4+ T lymphocytes in peripheral blood is a highly sensitive marker for early diagnosis and therapeutic monitoring of patients with Sezary syndrome. Clin Exp Dermatol 2005;30:702-6. [Crossref] [PubMed]
- Langerak AW, van Den Beemd R, Wolvers-Tettero IL, et al. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood 2001;98:165-73. [Crossref] [PubMed]
- Yawalkar N, Ferenczi K, Jones DA, et al. Profound loss of T-cell receptor repertoire complexity in cutaneous T-cell lymphoma. Blood 2003;102:4059-66. [Crossref] [PubMed]
- Kirsch IR, Watanabe R, O’Malley JT, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med 2015;7:308ra158. [Crossref] [PubMed]
- Dulmage B, Geskin L, Guitart J, et al. The biomarker landscape in mycosis fungoides and Sézary syndrome. Exp Dermatol 2017;26:668-76. [Crossref] [PubMed]
- Bensussan A, Remtoula N, Sivori S, et al. Expression and function of the natural cytotoxicity receptor NKp46 on circulating malignant CD4+ T lymphocytes of Sézary syndrome patients. J Invest Dermatol 2011;131:969-76. [Crossref] [PubMed]
- Nikolova M, Musette P, Bagot M, et al. Engagement of ILT2/CD85j in Sézary syndrome cells inhibits their CD3/TCR signaling. Blood 2002;100:1019-25. [Crossref] [PubMed]
- Bagot M, Moretta A, Sivori S, et al. CD4(+) cutaneous T-cell lymphoma cells express the p140-killer cell immunoglobulin-like receptor. Blood 2001;97:1388-91. [Crossref] [PubMed]
- Poszepczynska-Guigné E, Schiavon V, D’Incan M, et al. CD158k/KIR3DL2 Is a New Phenotypic Marker of Sezary Cells: Relevance for the Diagnosis and Follow-Up of Sezary Syndrome. J Invest Dermatol 2004;122:820-3. [Crossref] [PubMed]
- Nguyen GH, Olson LC, Magro CM. Upregulation of inhibitory signaling receptor programmed death marker-1 (PD-1) in disease evolution from cutaneous lymphoid dyscrasias to mycosis fungoides and Sezary’s syndrome. Ann Diagn Pathol 2017;28:54-9. [Crossref] [PubMed]
- Scala E, Abeni D, Pomponi D, et al. The role of 9-O-acetylated ganglioside D3 (CD60) and {alpha}4{beta}1 (CD49d) expression in predicting the survival of patients with Sezary syndrome. Haematologica 2010;95:1905-12. [Crossref] [PubMed]
- Chung J-S, Shiue LH, Duvic M, et al. Sézary syndrome cells overexpress syndecan-4 bearing distinct heparan sulfate moieties that suppress T-cell activation by binding DC-HIL and trapping TGF-beta on the cell surface. Blood 2011;117:3382-90. [Crossref] [PubMed]
- Wysocka M, Kossenkov AV, Benoit BM, et al. CD164 and FCRL3 Are Highly Expressed on CD4+CD26 − T Cells in Sézary Syndrome Patients. J Invest Dermatol 2014;134:229-36. [Crossref] [PubMed]
- Kamarashev J, Burg G, Kempf W, et al. Comparative analysis of histological and immunohistological features in mycosis fungoides and Sézary syndrome. J Cutan Pathol 1998;25:407-12. [Crossref] [PubMed]
- Huet D, Bagot M, Loyaux D, et al. SC5 mAb represents a unique tool for the detection of extracellular vimentin as a specific marker of Sezary cells. J Immunol 2006;176:652-9. [Crossref] [PubMed]
- Kari L, Loboda A, Nebozhyn M, et al. Classification and prediction of survival in patients with the leukemic phase of cutaneous T cell lymphoma. J Exp Med 2003;197:1477-88. [Crossref] [PubMed]
- Su MW, Dorocicz I, Dragowska WH, et al. Aberrant expression of T-plastin in Sezary cells. Cancer Res 2003;63:7122-7. [PubMed]
- Samaridis J, Colonna M. Cloning of novel immunoglobulin superfamily receptors expressed on human myeloid and lymphoid cells: Structural evidence for new stimulatory and inhibitory pathways. Eur J Immunol 1997;27:660-5. [Crossref] [PubMed]
- Chan AT, Kollnberger SD, Wedderburn LR, et al. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum 2005;52:3586-95. [Crossref] [PubMed]
- Carrillo-Bustamante P, Keşmir C, de Boer RJ. The evolution of natural killer cell receptors. Immunogenetics 2016;68:3-18. [Crossref] [PubMed]
- Bowness P, Ridley A, Shaw J, et al. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J Immunol 2011;186:2672-80. [Crossref] [PubMed]
- Fabre J, Giustiniani J, Garbar C, et al. Targeting the Tumor Microenvironment: The Protumor Effects of IL-17 Related to Cancer Type. Int J Mol Sci 2016;17. [Crossref] [PubMed]
- Bahler DW, Hartung L, Hill S, et al. CD158k/KIR3DL2 is a useful marker for identifying neoplastic T-cells in Sézary syndrome by flow cytometry. Cytometry B Clin Cytom 2008;74:156-62. [Crossref] [PubMed]
- Bouaziz JD, Remtoula N, Bensussan A, et al. Absolute CD3+ CD158k+ lymphocyte count is reliable and more sensitive than cytomorphology to evaluate blood tumour burden in Sézary syndrome. Br J Dermatol 2010;162:123-8. [Crossref] [PubMed]
- Moins-Teisserenc H, Daubord M, Clave E, et al. CD158k is a reliable marker for diagnosis of Sézary syndrome and reveals an unprecedented heterogeneity of circulating malignant cells. J Invest Dermatol 2015;135:247-57. [Crossref] [PubMed]
- Marie-Cardine A, Viaud N, Thonnart N, et al. IPH4102, a humanized KIR3DL2 antibody with potent activity against cutaneous T-cell lymphoma. Cancer Res 2014;74:6060-70. [Crossref] [PubMed]
- Ortonne N, Huet D, Gaudez C, et al. Significance of circulating T-cell clones in Sezary syndrome. Blood 2006;107:4030-8. [Crossref] [PubMed]
- Hurabielle C, Thonnart N, Ram-Wolff C, et al. Usefulness of KIR3DL2 to Diagnose, Follow-Up, and Manage the Treatment of Patients with Sézary Syndrome. Clin Cancer Res 2017;23:3619-27. [Crossref] [PubMed]
- Fife BT, Pauken KE. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann N Y Acad Sci 2011;1217:45-59. [Crossref] [PubMed]
- Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 2000;192:1027-34. [Crossref] [PubMed]
- Syn NL, Teng MWL, Mok TSK, et al. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol 2017;18:e731-41. [Crossref] [PubMed]
- Yu H, Shahsafaei A, Dorfman DM. Germinal-Center T-Helper-Cell Markers PD-1 and CXCL13 Are Both Expressed by Neoplastic Cells in Angioimmunoblastic T-Cell Lymphoma. Am J Clin Pathol 2009;131:33-41. [Crossref] [PubMed]
- Wada DA, Wilcox RA, Harrington SM, et al. Programmed death 1 is expressed in cutaneous infiltrates of mycosis fungoides and Sézary syndrome. Am J Hematol 2011;86:325-7. [Crossref] [PubMed]
- Samimi S, Benoit B, Evans K, et al. Increased programmed death-1 expression on CD4+ T cells in cutaneous T-cell lymphoma: implications for immune suppression. Arch Dermatol 2010;146:1382-8. [Crossref] [PubMed]
- Cetinözman F, Jansen PM, Vermeer MH, et al. Differential Expression of Programmed Death-1 (PD-1) in Sézary Syndrome and Mycosis Fungoides. Arch Dermatol 2012;148:1379-85. [Crossref] [PubMed]
- Wada T, Seki H, Konno A, et al. Developmental changes and functional properties of human memory T cell subpopulations defined by CD60 expression. Cell Immunol 1998;187:117-23. [Crossref] [PubMed]
- Kniep B, Kniep E, Ozkucur N, et al. 9-O-acetyl GD3 protects tumor cells from apoptosis. Int J Cancer 2006;119:67-73. [Crossref] [PubMed]
- Baciu PC, Saoncella S, Lee SH, et al. Syndesmos, a protein that interacts with the cytoplasmic domain of syndecan-4, mediates cell spreading and actin cytoskeletal organization. J Cell Sci 2000;113:315-24. [PubMed]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem 2002;71:435-71. [Crossref] [PubMed]
- Bühling F, Kunz D, Reinhold D, et al. Expression and functional role of dipeptidyl peptidase IV (CD26) on human natural killer cells. Nat Immun 1994;13:270-9. [PubMed]
- Ikushima H, Munakata Y, Ishii T, et al. Internalization of CD26 by mannose 6-phosphate/insulin-like growth factor II receptor contributes to T cell activation. Proc Natl Acad Sci U S A 2000;97:8439-44. [Crossref] [PubMed]
- Ludwig A, Schiemann F, Mentlein R, et al. Dipeptidyl peptidase IV (CD26) on T cells cleaves the CXC chemokine CXCL11 (I-TAC) and abolishes the stimulating but not the desensitizing potential of the chemokine. J Leukoc Biol 2002;72:183-91. [PubMed]
- Stanford MM, Issekutz TB. The relative activity of CXCR3 and CCR5 ligands in T lymphocyte migration: concordant and disparate activities in vitro and in vivo. J Leukoc Biol 2003;74:791-9. [Crossref] [PubMed]
- Watt SM, Bühring HJ, Rappold I, et al. CD164, a novel sialomucin on CD34(+) and erythroid subsets, is located on human chromosome 6q21. Blood 1998;92:849-66. [PubMed]
- Davis RS, Dennis G, Kubagawa H, et al. Fc receptor homologs (FcRH1-5) extend the Fc receptor family. Curr Top Microbiol Immunol 2002;266:85-112. [Crossref] [PubMed]
- Carreno BM, Bennett F, Chau TA, et al. CTLA-4 (CD152) can inhibit T cell activation by two different mechanisms depending on its level of cell surface expression. J Immunol 2000;165:1352-6. [Crossref] [PubMed]
- Wong HK, Wilson AJ, Gibson HM, et al. Increased Expression of CTLA-4 in Malignant T Cells from Patients with Mycosis Fungoides - Cutaneous T-Cell Lymphoma. J Invest Dermatol 2006;126:212-9. [Crossref] [PubMed]
- Gibson HM, Mishra A, Chan DV, et al. Impaired Proteasome Function Activates GATA3 in T Cells and Upregulates CTLA-4: Relevance for Sézary Syndrome. J Invest Dermatol 2013;133:249-57. [Crossref] [PubMed]
- Slavik JM, Hutchcroft JE, Bierer BE. CD28/CTLA-4 and CD80/CD86 families. Immunol Res 1999;19:1-24. [Crossref] [PubMed]
- Prasad A, Rabionet R, Espinet B, et al. Identification of Gene Mutations and Fusion Genes in Patients with Sézary Syndrome. J Invest Dermatol 2016;136:1490-9. [Crossref] [PubMed]
- Sekulic A, Liang WS, Tembe W, et al. Personalized treatment of Sézary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet genomic Med 2015;3:130-6. [Crossref] [PubMed]
- Dulmage BO, Geskin LJ. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. Br J Dermatol 2013;169:1188-97. [Crossref] [PubMed]
- Nebozhyn M, Loboda A, Kari L, et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood 2006;107:3189-96. [Crossref] [PubMed]
- Michel L, Jean-Louis F, Begue E, et al. Use of PLS3, Twist, CD158k/KIR3DL2, and NKp46 gene expression combination for reliable Sézary syndrome diagnosis. Blood 2013;121:1477-8. [Crossref] [PubMed]
- Litvinov IV, Netchiporouk E, Cordeiro B, et al. The Use of Transcriptional Profiling to Improve Personalized Diagnosis and Management of Cutaneous T-cell Lymphoma (CTCL). Clin Cancer Res 2015;21:2820-9. [Crossref] [PubMed]
- Boonk SE, Zoutman WH, Marie-Cardine A, et al. Evaluation of Immunophenotypic and Molecular Biomarkers for Sézary Syndrome Using Standard Operating Procedures: A Multicenter Study of 59 Patients. J Invest Dermatol 2016;136:1364-72. [Crossref] [PubMed]
- Boonk SE, Zoutman WH, Putter H, et al. Increased Expression of PLS3 Correlates with Better Outcome in Sézary Syndrome. J Invest Dermatol 2017;137:754-7. [Crossref] [PubMed]
- Caprini E, Cristofoletti C, Arcelli D, et al. Identification of key regions and genes important in the pathogenesis of Sézary syndrome by combining genomic and expression microarrays. Cancer Res 2009;69. [Crossref] [PubMed]
- Litvinov IV, Tetzlaff MT, Thibault P, et al. Gene expression analysis in Cutaneous T-Cell Lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology 2017;6:e1306618. [Crossref] [PubMed]
- Lefrançois P, Tetzlaff MT, Moreau L, et al. TruSeq-Based Gene Expression Analysis of Formalin-Fixed Paraffin-Embedded (FFPE) Cutaneous T-Cell Lymphoma Samples: Subgroup Analysis Results and Elucidation of Biases from FFPE Sample Processing on the TruSeq Platform. Front Med (Lausanne) 2017;4:153. [Crossref] [PubMed]
- Iżykowska K, Przybylski GK, Gand C, et al. Genetic rearrangements result in altered gene expression and novel fusion transcripts in Sézary syndrome. Oncotarget 2017;8:39627-39. [Crossref] [PubMed]
- Ververis K, Hiong A, Karagiannis TC, et al. Histone deacetylase inhibitors (HDACIs): multitargeted anticancer agents. Biologics 2013;7:47-60. [PubMed]
- Halsall JA, Turner BM. Histone deacetylase inhibitors for cancer therapy: An evolutionarily ancient resistance response may explain their limited success. Bioessays 2016;38:1102-10. [Crossref] [PubMed]
- Ralfkiaer U, Hagedorn PH, Bangsgaard N, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011;118:5891-900. [Crossref] [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [Crossref] [PubMed]
- Ballabio E, Mitchell T, van Kester MS, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood 2010;116:1105-13. [Crossref] [PubMed]
- Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 2004;101:2999-3004. [Crossref] [PubMed]
- Narducci MG, Arcelli D, Picchio MC, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis 2011;2. [Crossref] [PubMed]
- Cristofoletti C, Picchio MC, Lazzeri C, et al. Comprehensive analysis of PTEN status in Sezary syndrome. Blood 2013;122:3511-20. [Crossref] [PubMed]
- Cristofoletti C, Picchio MC, Russo G. MicroRNA-mediated Gene Expression in Sezary Syndrome: An Overview. Int Trends Immun 2015;3. [Crossref]
- Kopp KL, Ralfkiaer U, Mette Gjerdrum L, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell Cycle 2013;12:1939-47. [Crossref] [PubMed]
- Dusílková N, Bašová P, Polívka J, et al. Plasma miR-155, miR-203, and miR-205 are Biomarkers for Monitoring of Primary Cutaneous T-Cell Lymphomas. Int J Mol Sci 2017;18. [Crossref] [PubMed]
- Lee CS, Ungewickell A, Bhaduri A, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood 2012;120:3288-97. [Crossref] [PubMed]
- Kiel MJ, Sahasrabuddhe AA, Rolland DCM, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sézary syndrome. Nat Commun 2015;6:8470. [Crossref] [PubMed]
- Wang L, Ni X, Covington KR, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet 2015;47:1426-34. [Crossref] [PubMed]
- Vermeer MH, van Doorn R, Dijkman R, et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res 2008;68:2689-98. [Crossref] [PubMed]
- Mao X, Lillington D, Scarisbrick JJ, et al. Molecular cytogenetic analysis of cutaneous T-cell lymphomas: identification of common genetic alterations in Sézary syndrome and mycosis fungoides. Br J Dermatol 2002;147:464-75. [Crossref] [PubMed]
- Lin WM, Lewis JM, Filler RB, et al. Characterization of the DNA copy-number genome in the blood of cutaneous T-cell lymphoma patients. J Invest Dermatol 2012;132:188-97. [Crossref] [PubMed]
- Lamprecht B, Kreher S, Möbs M, et al. The tumour suppressor p53 is frequently nonfunctional in Sézary syndrome. Br J Dermatol 2012;167:240-6. [Crossref] [PubMed]
- Jones CL, Wain EM, Chu CC, et al. Downregulation of Fas gene expression in Sézary syndrome is associated with promoter hypermethylation. J Invest Dermatol 2010;130:1116-25. [Crossref] [PubMed]
- Laharanne E, Chevret E, Idrissi Y, et al. CDKN2A-CDKN2B deletion defines an aggressive subset of cutaneous T-cell lymphoma. Mod Pathol 2010;23:547-58. [Crossref] [PubMed]
- Steininger A, Möbs M, Ullmann R, et al. Genomic loss of the putative tumor suppressor gene E2A in human lymphoma. J Exp Med 2011;208:1585-93. [Crossref] [PubMed]
- Braun FCM, Grabarczyk P, Möbs M, et al. Tumor suppressor TNFAIP3 (A20) is frequently deleted in Sézary syndrome. Leukemia 2011;25:1494-501. [Crossref] [PubMed]
- McGirt LY, Jia P, Baerenwald DA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood 2015;126:508-19. [Crossref] [PubMed]
- Choi J, Goh G, Walradt T, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet 2015;47:1011-9. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Bastidas Torres AN, Najidh S, Tensen CP, et al. Molecular advances in cutaneous T-cell lymphoma. Semin Cutan Med Surg 2018;37:81-6. [Crossref] [PubMed]
- Woollard WJ, Pullabhatla V, Lorenc A, et al. Candidate driver genes involved in genome maintenance and DNA repair in Sézary syndrome. Blood 2016;127:3387-97. [Crossref] [PubMed]
- Ungewickell A, Bhaduri A, Rios E, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat Genet 2015;47:1056-60. [Crossref] [PubMed]
- da Silva Almeida AC, Abate F, Khiabanian H, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat Genet 2015;47:1465-70. [Crossref] [PubMed]
- Karenko L, Hahtola S, Päivinen S, et al. Primary cutaneous T-cell lymphomas show a deletion or translocation affecting NAV3, the human UNC-53 homologue. Cancer Res 2005;65:8101-10. [Crossref] [PubMed]
- Vaqué JP, Gómez-López G, Monsálvez V, et al. PLCG1 mutations in cutaneous T-cell lymphomas. Blood 2014;123:2034-43. [Crossref] [PubMed]
- Park J, Yang J, Wenzel AT, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood 2017;130:1430-40. [Crossref] [PubMed]
- Annala MJ, Parker BC, Zhang W, et al. Fusion genes and their discovery using high throughput sequencing. Cancer Lett 2013;340:192-200. [Crossref] [PubMed]
- Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med 1995;182:459-65. [Crossref] [PubMed]
- Chase A, Ernst T, Fiebig A, et al. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica 2010;95:20-6. [Crossref] [PubMed]
- Talpur R, Bassett R, Duvic M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sézary syndrome. Br J Dermatol 2008;159:105-12. [Crossref] [PubMed]
- Trautinger F, Eder J, Assaf C, et al. European Organisation for Research and Treatment of Cancer consensus recommendations for the treatment of mycosis fungoides/Sézary syndrome - Update 2017. Eur J Cancer 2017;77:57-74. [Crossref] [PubMed]
- Zelenetz AD, Gordon LI, Wierda WG, et al. Non-Hodgkin’s lymphomas, version 4.2014. J Natl Compr Canc Netw 2014;12:1282-303. [Crossref] [PubMed]
- Virmani P, Hwang SH, Hastings JG, et al. Systemic therapy for cutaneous T-cell lymphoma: who, when, what, and why? Expert Rev Hematol 2017;10:111-21. [Crossref] [PubMed]
- Molina A, Zain J, Arber DA, et al. Durable clinical, cytogenetic, and molecular remissions after allogeneic hematopoietic cell transplantation for refractory Sezary syndrome and mycosis fungoides. J Clin Oncol 2005;23:6163-71. [Crossref] [PubMed]
- Sors A, Jean-Louis F, Pellet C, et al. Down-regulating constitutive activation of the NF-kappaB canonical pathway overcomes the resistance of cutaneous T-cell lymphoma to apoptosis. Blood 2006;107:2354-63. [Crossref] [PubMed]
- Nicolay JP, Müller-Decker K, Schroeder A, et al. Dimethyl fumarate restores apoptosis sensitivity and inhibits tumor growth and metastasis in CTCL by targeting NF-κB. Blood 2016;128:805-15. [Crossref] [PubMed]
- Zinzani PL, Musuraca G, Tani M, et al. Phase II trial of proteasome inhibitor bortezomib in patients with relapsed or refractory cutaneous T-cell lymphoma. J Clin Oncol 2007;25:4293-7. [Crossref] [PubMed]
- Olsen EA, Kim YH, Kuzel TM, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol 2007;25:3109-15. [Crossref] [PubMed]



