Inherited pancreatic cancer
Family history and risk of pancreatic cancer
Family history is a long-recognized risk factor for pancreatic cancer and an important predictor of disease risk. Studies have suggested that approximately 5–10% of pancreatic cancer patients report a close relative with pancreatic cancer (1,2). Most epidemiological studies have demonstrated a 2- to 3-fold increase in risk of pancreatic cancer among individual with affected first-degree relatives (FDRs) (3-12). However, some studies have shown even higher risk. A Swedish study reported a standardized incidence ratios (SIRs) for pancreatic cancer of 1.73 (95% CI: 1.13–2.54) in offspring with at least one parent presented with pancreatic ductal adenocarcinoma (PDAC) (13). In a prospective study from the National Familial Pancreas Tumour Registry (NFPTR), the SIRs for pancreatic cancer in comparison to the Surveillance, Epidemiology, and End Results (SEER) rates were 6.4 (95% CI: 1.8–16.4) and 32.0 (95% CI: 10.2–74.7) in individuals with two and three FDRs with pancreatic cancer (14).
Familial clustering was considered the first evidence supporting the genetic predisposition to pancreatic cancer. Reports of multiple siblings in one generation and individuals in three consecutive generations affected by pancreatic cancer are strong evidence of a hereditary form of the disease following the Mendelian inheritance (15-19). This was later supported by segregation analyses, which favored a major gene model that was predicted to follow an autosomal dominant pattern of a rare allele (20). As demonstrated in the observational epidemiologic studies, individuals with a family history of pancreatic cancer are at an increased risk of developing the disease themselves. In addition, a population-based twin study in Europe has estimated the heritability for pancreatic cancer to be 36% (95% CI: 0–53%) (21). As more evidence of the genetic basis of pancreatic cancer has emerged, an operational definition of familial pancreatic cancer (FPC) was proposed to facilitate investigations of the inherited components of the disease. FPC is defined as kindreds with at least a pair of FDRs diagnosed with PDAC. Comparing to the general population, FPC kindred members have a 7- to 9-fold increased risk of pancreatic cancer (14,22). Risk is even higher among members of FPC kindreds with a young-onset case (<50 years; SIR =9.31; 95% CI: 3.42–20.28) than those without (SIR =6.34; 95% CI: 4.02–9.51) (22).
Both prospective and retrospective studies have found increased risks of other cancers in relatives of pancreatic cancer patients, particularly breast cancer (23,24), melanoma (24), ovarian (25), and colorectal cancer (24,26). Study from NFPTR reported an increased risk of dying from cancer of the breast [weighted standardized mortality ratio (wSMR) =1.66; 95% CI: 1.15–2.34], ovarian (wSMR= 2.05; 95% CI: 1.10–3.49), bile duct (wSMR =2.89; 95% CI: 1.04–6.39) and bladder (wSMR =1.90; 95% CI: 1.00–3.30) in FDRs of FPC probands (27). Elevated mortality of colon cancer (wSMR=2.31; 95% CI: 1.30–3.81) and prostate cancer (wSMR=2.31; 95% CI: 1.14–4.20) were also observed among the relatives of young-onset (<50 years old) pancreatic cancer probands (27). These findings suggest a shared genetic etiology between pancreatic cancer and several other cancers, and the potential benefits of surveillance at-risk relatives for cancers with established screening guidelines.
Pancreatic cancer genes
Pancreatic cancer is more prevalent in families with several hereditary syndromes, for which the predisposing genes have been identified, including BRCA1 and BRCA2 associated with hereditary breast and ovarian cancer (HBOC), STK11 associated with Peutz-Jeghers syndrome (PJS), CDKN2A/p16 associated with familial atypical mole and multiple melanoma (FAMMM), mismatch repair (MMR) genes associated with Lynch syndrome, and PRSS1 associated with hereditary pancreatitis (HP). These genetic syndromes are reported to be associated with a substantially higher risk of pancreatic cancer. The recent discovery of germline mutations in PALB2 and ATM gene in FPC kindreds has extended the list of established high- and moderate-risk pancreatic cancer genes (Table 1).
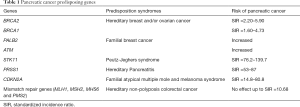
Full table
BRCA1/2
BRCA1 and BRCA2 gene are well-known high-penetrant predisposing genes for HBOC. These genes are involved in the DNA damage response and DNA double-strand breaks repair. Pancreatic cancer is the third most common cancer associated with BRCA1/2 mutations, though the penetrance at age 70 is much lower (28-31). The prevalence of BRCA2 mutations in pancreatic cancer patients is 1.4–8.2% for patients unselected for family history (32-36), about 6–16% among FPC patients (37-42), and up to 17.2% in families with 3 or more pancreatic cancers (38,41,43). Comparing to the general population, the risk of pancreatic cancer is about 2–6 fold in BRCA2 carriers (28,44) and 2–5 fold in BRCA1 carriers (29,30,44). Several studies had reported a higher risk of pancreatic cancer in BRCA2 carriers than in BRCA1 carriers (28,31,44-46). BRCA1/2 mutation carriers are at particularly high risk (SIR =4–10) for early onset pancreatic cancer (28-30,44). FDRs of BRCA1/2 carriers, regardless of their carrier status, have a significantly higher risk of pancreatic cancer than the general population (44,46).
PALB2
PALB2 gene is a tumor suppressor that interacts closely with both BRCA1 and BRCA2 during double-strand DNA repair. Mutations of PALB2 had previously been associated with familial breast cancer (47). Jones et al. first reported the discovery of truncating mutations of PALB2 gene in four FPC probands from the NFPTR (48). Since then, pathogenic mutations of PALB2 gene have been found in 0.4–4% FPC families, majority of which were families with history of both pancreatic cancer and breast/ovarian cancer (49-55).
ATM
ATM is a breast cancer susceptibility gene that coordinates the DNA double-strand breaks repair. Deleterious mutations of ATM gene were first reported by Roberts and his colleagues in two FPC families with at least three members affected by pancreatic cancer (56). In the subsequent analysis, four additional ATM mutations were found in 166 FPC patients compared to none in 190 spouse controls (56). To date, ATM mutations are found in 1–5% patients with pancreatic cancer (35,36,42,57-59).
STK11
PJS is caused by germline mutations in the STK11 gene (60-62). PJS patients are at very high risk of developing cancer during their lifetimes, particularly gastrointestinal cancer and gynecological cancer. The cumulative risk of developing any gastrointestinal cancer is 38–66% at age 70 (63). Compared to the general population, PJS patients have a 76- to 140-fold elevated risk for pancreatic cancer (64-66). The cumulative risk of developing pancreatic cancer at age 70 in PJS patients is 11–55% (64-67).
CDKN2A
CDKN2A is a tumor suppressor gene that is considered a major cause of familial melanoma. In melanoma-prone families of European ancestry, pancreatic cancer is the second most common type of cancers associated with CDKN2A mutations. Longitudinal studies in these families have found a 15- to 80-fold increased risk of pancreatic cancer in carriers of CDKN2A mutations comparing to the general population (68-73). The risk of developing pancreatic cancer is also higher in FDRs of carriers than in FDRs of non-carriers (RR =7.4; 95% CI: 2.3–18.7) (74).
MMR genes
Hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch Syndrome, accounts for 2–5% of all colorectal cancer. It is caused by inactivating mutations of DNA MMR genes: MLH1, MSH2, MSH6 and PMS2. While several studies found no increase in risk of pancreatic cancer in Lynch Syndrome patients (75-78), others reported an approximately 7- to 10-fold elevated risk of developing pancreatic cancer in carriers of MMR gene mutations (79-82). The relative risk of pancreatic cancer is higher at younger age (79,80). The cumulative risk of developing pancreatic cancer at age 70 among mutation carriers was estimated to be 3.68% (95% CI: 1.45–5.88%) (80).
PRSS1
HP is autosomal-dominant disorder characterized by recurrent episodes of acute pancreatitis in childhood and frequent progression to chronic pancreatitis. Germline mutations in PRSS1 are responsible for the majority of HP cases. Comparing to the general population, the risk of pancreatic cancer is about 69-fold higher in HP patients, and the median age of cancer onset was at least 15 years earlier (83-86). About 20–50% HP patients would develop pancreatic cancer at age 70 (83-85,87). The risk is even higher among smokers with HP who tend to develop pancreatic cancer 20 years before non-smokers (85,88).
Germline mutations in sporadic cases
Inherited genetic alterations are not restricted to patients with FDRs affected by pancreatic cancer. While current guidelines recommend germline genetic testing for pancreatic cancer patients with a first degree relative with pancreatic cancer or pancreatic cancer patients with a family history indicative of one of the above mentioned genetic syndromes, patients with apparently sporadic pancreatic cancer may also harbor mutation in a pancreatic cancer susceptibility gene. In fact, the discovery of the role of BRCA2 in pancreatic cancer was based upon the observation of Three germline mutations in BRCA2 was found in 41 (7.3%) sporadic pancreatic cancer patients (32). Subsequent studies have showed that in a series of 306 unselected PDAC patients, 14 carried mutations in BRCA1 or BRCA2 while only 2 of the 14 had a family history of PDAC (33). Salo-Mullen et al. reported a 7.4% prevalence of BRCA mutations in 27 PDAC patients of Ashkenazi Jewish ancestry without a family history of breast, ovarian or pancreatic cancer (34). Studies have reported 0–3% sporadic or unselected pancreatic cancer patients carrying PALB2 mutations, leading to an aggregated prevalence of 0.75% (55). Recently, in an evaluation of 854 pancreatic cancer patients, twelve of them had germline BRCA2 mutations, ten with ATM, three with BRCA1, and two with PALB2 (36). Larger scale studies are currently underway to evaluate the mutation prevalence in apparently sporadic pancreatic cancer and expanding genetic testing beyond the current guidelines.
Targeted therapy
Understanding genetic predisposition of pancreatic cancer has important implication for the development and translation of targeted therapies. Tumors with mutations in BRCA1/2, PALB2, and ATM are highly sensitive to DNA-damaging related treatments such as crosslinking agents (89-91) and poly(ADP-ribose) polymerase inhibitors (PARPi) (92-95). Preclinical studies have demonstrated improved sensitivity to chemotherapeutic agents and ionizing radiation in pancreatic cancer cells treated by these agents (96-98). The clinical benefits of using crosslinking agents and PARPi in patients with pancreatic cancer are currently being investigated. Preliminary results have shown promising efficacy of these agents, particularly in patients with BRCA2-associated pancreatic cancer (99-102).
Common low-risk susceptibility loci
Genome-wide association studies (GWAS) allow for the unbiased evaluation of common genomic variants associated with pancreatic cancer. To identify common susceptibility variants, five pancreatic cancer GWAS have been conducted by the Pancreatic Cancer Cohort Consortium (PanScan) and the Pancreatic Cancer Case Control Consortium (PanC4) in populations of European ancestry, including PanScan I in 2009 (103), PanScan II in 2010 (104), PanScan III in 2014 (105), PanC4 in 2015 (106) and a recent imputation analysis of GWAS data from PanScan I–III (107). A total of 16 pancreatic cancer susceptibility loci located in 13 genomic regions have been discovered in European populations (Table 2).
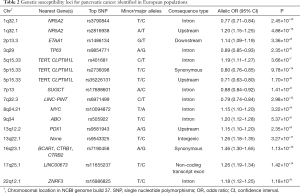
Full table
PanScan I
PanScan I was a two-stage GWAS including 1,896 patients with incidence pancreatic cancer and 1,939 controls in the discovery stage, as well as 2,457 cases and 2,654 controls in the replication stage. The most significant variant (rs505922) on chromosome 9q34.2 was mapped to the first intron of ABO blood group gene (103). The association of ABO loci with pancreatic cancer has been robustly replicated in studies of European (105,106,108,109) and Asian populations (110-112). These findings are consistent with the observation that individuals with blood group O had a lower risk of pancreatic cancer than those with groups A or B. About 17% to 19.5% of all pancreatic cancers in populations of European descent was attributable to the inheritance of a non-O blood group (113,114).
PanScan II
From 3,851 pancreatic cancer cases and 3,934 controls, PanScan II identified three novel genomic regions on chromosome 13q22.1 (a large non-genic region), chromosome 1q32.1 (NR5A2) and chromosome 5p15.33 (TERT-CLPTM1L) to be significantly associated with pancreatic cancer (104). The locus on chromosome 13q22.1 (rs9543325) was mapped to a large gene desert flanked by the KLF5 and KLF12 genes. Both genes encode a transcription factor involved in cell transformation, proliferation, and carcinogenesis. Several studies have reported the overexpression of the KLF5 gene in pancreatic cancer (115-118). The KLF2 gene, on the other hand, was found to be downregulated in PDAC tumor tissues, and its expression may suppress the malignant transformation of PDAC cancer cells through its regulation of beta-catenin/TCF signaling (119).
Two variants on chromosome 1q32.1 are associated with pancreatic cancer independently. The first significant variant (rs3790844) identified in PanScan II is located in the first intron of the NR5A2 gene (104). Imputation analysis of PanScan I–III detected the second variant in the upstream of NR5A2 (rs2816938). NR5A2 encodes the nuclear receptor subfamily 5 group A member 2, a transcription factor that activates or inhibits transcription of specific target genes. Overexpression of NR5A2 was observed in resected PDAC tumor tissues and was associated with reduced survival time in PDAC patients (120). Heterozygous Nr5a2 mice exhibit increased rates of pancreatic acinar to ductal metaplasia and impaired recovery after chemically induced acute pancreatitis (121,122). Loss of Nr5a2 accelerates the development of oncogenesis driven by Kras (121,122). These findings suggest a tumor suppressor role of NR5A2 that protects the pancreas from KRAS driven pre-neoplastic changes.
Four independent pancreatic cancer risk loci have now been identified in the multi-cancer TERT-CLPTM1L region on chromosome 5p15.33. The first pancreatic cancer risk locus identified in PanScan II is located in the intron 13 of CLPTM1L (rs401681). PanScan III reported a second independent risk locus on chromosome 5p15.33, tagged by a synonymous variant within the second exon of TERT (rs2736098) (105). A third independent risk locus located in the first intron of TERT gene (rs2853677) was discovered through a candidate gene analysis in 5,550 pancreatic cancer cases and 7,585 control subjects from PANDoRA (PANcreatic Disease ReseArch) consortium and PanScan (123). Recently, imputation of PanScan I–III and replication in PANDoRA and PanC4 found a fourth risk locus for pancreatic cancer in this genomic region (rs35226131), which is located about 200bps upstream of the transcriptional start site of TERT (107). The chromosome 5p15.33 region contains two plausible candidate genes: TERT, which encodes the catalytic subunit of telomerase reverse transcriptase and CLPTM1L, which encodes the cleft lip and palate-associated transmembrane 1 like protein. TERT is a component of the protein and RNA complex that maintains telomere ends. Mutations in TERT promoter region were frequent in multiple tumor types and were correlated with increased TERT expression and telomerase activation (124). Common variants in the TERT region were associated with leukocyte telomere length in patients with breast and ovarian cancer (125). A recent study had reported an association between the minor allele of rs401681 and shorter telomere length in pancreatic cancer patients, which was consistent with the observation that telomere shortening occurs as an early event in pancreatic tumorigenesis (126-128). Overexpression of CLPTM1L gene are observed in lung and pancreatic cancer tissues (129-131). CLPTM1L protects tumor cells from genotoxic apoptosis and is required for Ras-induced oncogenic transformation (129,130,132). It’s also found that overexpression of CLPTM1 may lead to an abrogation of normal cytokinesis and promote cell proliferation in pancreatic cancer cells (131).
PanScan III
The PanScan III study population combined 1,582 newly genotyped pancreatic cancer cases and 5,203 control subjects with PanScan I cohort and sought replication in 2,576 cases and 6,662 controls from the PANDoRA consortium. Four new risk loci for pancreatic cancer was identified on chromosome 7q23.2 (LINC-PINT), 16q23.1 (BCAR1), 13q12.2 (PDX1) and 22q12.1 (ZNRF3). The signal on 7q32.3 was marked by an intronic variant (rs6971499) in LINC-PINT, a long intergenic p53-induced non-protein coding RNA located between muskelin 1 (MKLN1) and Kruppel-like factor 14 (KLF14). Muskelin is an intracellular protein that mediates cell adhesive and cytoskeletal responses to the extracellular matrix (133). KLF14 is a member of the Kruppel-like family of transcription factors that may act as a suppressor of KRAS-mediated cell growth through regulation of the cyclin A promoter (134). Loss of KLF14 may also trigger centrosome amplification, aneuploidy and spontaneous tumorigenesis (135). KLF14 has also been associated with several metabolic phenotypes including type-2 diabetes mellitus (T2DM), a known risk factor for pancreatic cancer (136-139).
A synonymous variant residing in the last exon of BCAR1 was noted on 16q23.1 (rs7190458). Breast cancer anti-estrogen resistance 1 (BCAR1), also known as p130Cas is a member of the Cas (Crk-associated substrate) family of adaptor proteins with important regulatory roles in migration, cell cycle control and apoptosis (140). Altered expression and activity of p130Cas is known to promote metastasis and drug resistance in multiple cancers (140,141). In addition, two chymotrypsinogen genes, CTRB1 and CTRB2 are also located closely to the detected signal. As important members of a family of serine proteases secreted by the pancreas into the gastrointestinal tract (142), these two genes are plausible target for susceptibility variants at this locus. The detected signal for pancreatic cancer is also in proximity of a susceptibility locus (rs7202877) for type-I and type-II diabetes (143,144) that was found to impair beta-cell function (145) and influences expression of CTRB1/2 in pancreas tissues (146).
The top ranked variant on chromosome 13q12.2 (rs9581943) is located in the promoter region of the PDX1 (pancreatic and duodenal homeobox1 protein 1) gene. Pathway analysis of GWAS data identified PDX1, along with NR5A2, HNF1A, and HNF4G, as important genes for pancreatic development (147). The protein encoded by PDX1 is a transcriptional activator of several genes. It is essential in the early development of pancreas (148), and plays a major role in beta-cell function and glucose-dependent regulation of insulin gene expression. Heterozygous mutations in PDX1 resulted in impaired glucose tolerance and symptoms of diabetes as seen in maturity-onset diabetes of the young type 4 (MODY4) and late-onset T2DM (149-151).
The signal on chromosome 22q2.1 (rs16986825) maps to an intron in ZNRF3 (zinc and ring finger 3), which encodes a cell surface transmembrane E3 ubiquitin protein ligase that is a negative regulator of the WNT signaling pathway (152). Additionally, a low-penetrance breast cancer gene, CHEK2, is also located in proximity to the detected signal. This gene encodes a cell-cycle checkpoint kinase that cooperates with p53, BRCA1 and ATM and regulates cell division in response to DNA damage (153). Germline mutations and variants of CHEK2 had been implicated in susceptibility to several cancer types (154-158), including FPC (41,42,159).
PanC4
PanC4 was conducted on 9,925 pancreatic cancer cases and 11,569 controls, pooling 4,164 newly genotyped cases and 3,792 controls from nine studies in the PanC4 consortium with PanSan I and II cohorts in the gene discovery stage, and analyzed an independent set of 2,497 cases and 4,611 controls from the PANDoRA consortium in the replication stage. Not only this study replicated all previously identified risk loci for pancreatic cancer in European populations, three novel associated signals were also detected on chromosome 17q25.1 (LINC00673), 7p13 (SUGCT) and 3q29 (TP63) (106). Significant association was also found on chromosome 2q13.3 (ETAA1), a region with prior suggestive evidence in the Han Chinese (160).
The top variant on 17q25.1 (rs11655237) maps to LINC00673 (long inter-genic non-protein coding RNA 673). This association subsequently replicated in a Han Chinese population (rs11655237, OR=1.26, P=3.95×10‒14) (161). Through its epigenetic regulation of gene expression, LINC00673 may function as an oncogene in several types of cancers. Overexpression of LINC00673 promotes tumor proliferation, invasion and metastasis in non-small-cell lung cancer (162-164) and tongue squamous cell carcinoma (165), and was correlated with poor prognosis in breast cancer (166). In contrast, expression of LINC00673 was significantly lower in PDAC cancer cells than in normal cells and tissues and overexpression of LINC00673 in the PDAC cell line substantially reduced the rate of cell proliferation. It was found that the single-nucleotide change at rs11655237 creates a miR-1231 binding site, which diminishes the effect of LINC00673 in an allele-specific manner and thus confer susceptibility to pancreatic tumorigenesis (161).
PanC4 reported a significant association on 7p13 with an intronic variant (rs17688601) of the succinyl-CoA:glutarate-CoA transferase (SUGCT) gene. Mutations of this gene cause a benign form of glutaric aciduria (glutaric aciduria type III), a rare metabolic abnormality characterized by persistent isolated accumulation or excretion of glutaric acid (167). The role of this gene in pancreatic cancer risk is unclear.
Two strongly correlated intronic variants of TP63 (tumor protein p63) were found to be associated with pancreatic cancer in PanC4 (top variants rs9864771). Protein encoded by TP63 (p51/p63) is a p53 homologue with pleiotropic functions including cell proliferation, survival, apoptosis, differentiation, senescence, and aging. Frequent overexpression of p63 was observed in resected PDAC tissues (168). It was suggested that different isoforms of p63 have opposite effects. While TAp63 induces cell death and cell cycle arrest with tumor suppressor features (169), DNp63 as the predominant isoform in pancreatic cancer cell lines, promotes pancreatic cancer growth, motility and invasion (170,171). As a tumor suppressor, p63 had reduced anti-oncogenetic effects compared with p53 in human cancer cells (172). However, loss of p63 can cooperate with loss of p53, leading to higher tumor burden and metastasis as seen in genetic mice models (168,171). It is hypothesized that it is the ratio of TAp63 and DNp63 that determines the biological outcome and chemo-sensitivity.
Imputation analysis of PanScan I–III
Recently, an imputation analysis of the GWAS data in 5,107 cases and 8,845 controls from PanScan I–III had uncovered three new pancreatic cancer signals on chromosome 1q32.1 (NR5A2), 8q24.21 (MYC), and 5p15.33 (CLPTM1L-TERT), all of which are independent from previously reported susceptibility variants (107).
The detected variants on 8q24.21 (rs10094872) is a novel risk loci for pancreatic cancer, independent from the previously reported loci with suggestive evidence in PanScan III (rs1561927). These two variants are both located in the 2 Mb region known to contain multiple susceptibility loci that influence risk of bladder, breast, prostate, colorectal, lung, ovarian, pancreatic, and renal cancer (173-177). MYC (MYC proto-oncogene, bHLH transcription factor) is the gene located in the closest proximity to the detected variant. Oncogene MYC is a transcription factor that has been implicated in the pathogenesis of one-third of all human malignancies, and may play an important role in KRAS-driven neoplastic transformation in the pancreas (178). MYC overexpression occurs in up to 42% of advanced PDAC (179,180). Activation of MYC in adult mice has led to the development of ductal adenocarcinomas with metastasis to the liver (181). Although evidence have suggested regulatory roles of the 8q24.21 risk loci in the expression of MYC, functional analyses are warranted to allow a deeper understanding of the underlying mechanism (178).
GWAS in Asian populations
Two GWAS have been conducted in populations of Asian descent (Table 3). The Japanese pancreatic cancer study of 991 cases and 5,209 controls found suggestive associations on chromosome 6q25.3 (FOXQ1), 12p11.21 (BICD1) and 7q36.2 (DPP6) (182). The second GWAS in a Chinese population of 3,584 pancreatic cancer cases and 4,868 controls (ChinaPC) identified five susceptibility loci on chromosome 21q21.3 (BACH1), 21q22.3 (TFF1), 10q26.11 (PRLHR), 22q13.32 (FAM19A5), and 5p13.1 (DAB2) (160). The most significant association identified in ChinaPC was for rs372883, a variant located in the 3’ untranslated region (3’UTR) of BACH1 (BTB domain and CNC homolog 1) gene on chromosome 21q21.3. BACH1 is a transcription factor that belongs to the cap ‘n’ collar type of basic region leucine zipper factor family (CNC-bZip). Recent studies have demonstrated a critical role of BACH1 in cell migration and metastasis through its regulation of metastasis-related gene expression in breast, colon and prostate cancer (183-185). The second significant association was detected on chromosome 21q22.3 (rs1547374). This region harbors the trefoil family protein 1 (TFF1) gene that encodes secretory proteins expression in gastrointestinal mucosa. Upregulated expression of TFF1 in precursor lesions of PDAC, including pancreatic intraepithelial neoplasia (PanIN), intraductal papillary-mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MSNs), suggests its potential involvement at the early stage of pancreatic carcinogenesis (186-188). Recent studies found that reduced expression of TFF1 in the invasion front of human PDAC was associated with lymph node metastasis and poor survival in patients with PDAC (189). In pancreatic cancers, expression of TFF1 promotes tumorigenesis by suppressing oncogene-induced senescence (190) and is correlated with increase metastasis (191). An intronic variant (rs2255280) in DAB2 (clathrin adaptor protein) gene region on 5p13.1 was among the identified susceptibility loci in ChinaPC. Frequent loss expression of DAB2 in human malignant cancer cells suggests its potential role as a tumor suppressor (192). Overexpression of DAB2 inhibits cell growth, migration and invasion, and was correlated with poor survival in cancer patients (193-196). Significant association on 10q26.11 was observed for a regulatory variant of PRLHR (prolactin releasing hormone receptor) gene (rs12413624). Polymorphisms of this gene was associated with colorectal cancer (197). A intronic variant of FAM19A5 (family with sequence similarity 19 member A5) gene on 22q13.32 was also associated with an increased risk of pancreatic cancer (rs5768709). This gene encodes a TAFA protein expressed predominately in brain and may function as brain-specific chemokines or neurokines (198). Prior to the ChinaPC study, there is no implication of PRLHR or FAM19A5 in the risk of pancreatic cancer and thus their susceptibility role is currently unknown.

Full table
Current status of screening high-risk populations
Screening and early detection of pancreatic cancer offer the best chance of reducing the high mortality rates of this disease. The goal of screening asymptomatic individuals is to identify pancreatic cancer at early stage or, ideally to identify high-grade precancerous lesions that can be resected to prevent the development of cancer. Because of the low incidence of pancreatic cancer in the general population, population level screening will demand a highly specific screening assay. Selective screening of individuals at increased risk for pancreatic cancer is considered worthwhile. The International Cancer of the Pancreas Screening (CAPS) Consortium recommends screening on FDRs of FPC patients, patients with PJS, and carriers of CDKN2A/p16, BRCA2, and carriers of MMR gene mutations with ≥1 affected FDR (199). FDRs of FPC patients represent a group of high-risk individuals that are relatively easy to identify in clinical settings. Of all identified risk factors for pancreatic cancer, PJS confers the greatest risk for the disease, making PJS patients good candidates for pancreatic cancer screening. Among the established pancreatic cancer genes, germline BRCA2 mutations followed by ATM account for the highest percentage of inherited pancreatic cancer (34,36,40,42,58). It is recommended that BRCA2 mutation carriers with ≥1 affected FDR and those with two or more affected family members should be considered for screening, particularly Ashkenazi Jewish individuals. In addition, given the substantially higher risk of pancreatic cancer in patients with CDKN2A/p16 and MMR gene mutations, screening is also recommended to those mutation carriers with >1 affected FDRs (199).
Endoscopic ultrasonography (EUS) and/or MRI/magnetic resonance cholangiopancreatography (MRCP) are recommended by CAPS as initial screening tools. However, the CAPS Consortium could not reach consensus on ages to initiate or stop surveillance, the interval for follow-up imaging, nor on the long-term management of initial abnormal results (199). Screening and early detection strategies should be accompanied by effective treatment or preventive strategies if they are to produce a significant survival benefit. Given morbidity and mortality associated with pancreatic surgery, there is little consensus about when surgery is required for pancreatic lesion in asymptomatic high-risk individuals (199). Multidisciplinary assessment is however recommended to make individualized decision of the necessity of surgical intervention. The lack of consensus on many aspects of pancreatic cancer screening underscores the need for more research to fill the knowledge gap and to make evidence-based decisions.
Summary and future directions
Pancreatic cancer is rare and deadly disease with the highest case-fatality rate of any major cancer. Due to the lack of effective means for prevention, diagnosis and treatment, pancreatic cancer remains a major public health challenge. Family history, cigarette smoking, chronic pancreatitis, and diabetes are well-established risk factors for pancreatic cancer. Pancreatic cancer is fundamentally a genetic disease caused by both inherited and acquired genetic mutations. Family-based heritability analysis reported 36% of pancreatic cancer was due to genetics. FPC kindreds and patients affected by certain genetic syndromes, for example HP, PJS, HBOC, FAMMM, and HNPCC, are at particularly high risk of pancreatic cancer. About 15–20% of FPC are caused by germline mutations in one of the established pancreatic cancer gene (BRCA2, ATM, PALB2, PRSS1, STK11, BRCA1, CDKN2A, MLH1, MSH2, MSH6, and PMS2). The genetic basis of susceptibility underlying the majority of FPC cases, however, remains unexplained. To date, GWAS of pancreatic cancer have discovered 16 low-risk susceptibility loci in European populations and 5 in Asian populations, many of which had strong biological plausibility. Together, these GWAS loci explained <5% of pancreatic cancer. Screening, surveillance and management guidelines for genetically high-risk individuals are currently evolving. DNA-damaging related agents are promising in treating pancreatic cancer caused by mutations in BRCA1, BRCA2, PALB2 or ATM genes. Identification of disease-causing genes can aid in the characterization of individuals at highest genetically defined risk in which effective prevention approach can be developed.
Acknowledgements
Funding: This work is supported the NIH Specialized Programs of Research Excellence P50-CA062924, NIH grants R00-CA190889 and R01-CA154823 and the Lustgarten Foundation for Pancreatic Cancer Research and the Sol Goldman Pancreatic Cancer Research Center and the Burroughs-Wellcome Fund Maryland: Genetics, Epidemiology and Medicine Training Grant.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hruban RH, Petersen GM, Ha PK, et al. Genetics of pancreatic cancer. From genes to families. Surg Oncol Clin N Am 1998;7:1-23. [PubMed]
- Klein AP. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer 2013;13:66. [Crossref] [PubMed]
- Fernandez E, La Vecchia C, D’Avanzo B, et al. Family history and the risk of liver, gallbladder, and pancreatic cancer. Cancer Epidemiol Biomarkers Prev 1994;3:209-12. [PubMed]
- Silverman DT, Schiffman M, Everhart J, et al. Diabetes mellitus, other medical conditions and familial history of cancer as risk factors for pancreatic cancer. Br J Cancer 1999;80:1830-7. [Crossref] [PubMed]
- Schenk M, Schwartz AG, O’Neal E, et al. Familial risk of pancreatic cancer. J Natl Cancer Inst 2001;93:640-4. [Crossref] [PubMed]
- Fehringer G, Gallinger S, Borgida A, et al. The association of family history of cancer and medical history with pancreatic cancer risk. Pancreas 2014;43:812-4. [Crossref] [PubMed]
- Schulte A, Pandeya N, Fawcett J, et al. Association between family cancer history and risk of pancreatic cancer. Cancer Epidemiol 2016;45:145-50. [Crossref] [PubMed]
- Zheng Z, Zheng R, He Y, et al. Risk factors for pancreatic cancer in china: a multicenter case-control study. J Epidemiol 2016;26:64-70. [Crossref] [PubMed]
- Jacobs EJ, Rodriguez C, Newton CC, et al. Family history of various cancers and pancreatic cancer mortality in a large cohort. Cancer Causes Control 2009;20:1261-9. [Crossref] [PubMed]
- Permuth-Wey J, Egan KM. Family history is a significant risk factor for pancreatic cancer: results from a systematic review and meta-analysis. Fam Cancer 2009;8:109-17. [Crossref] [PubMed]
- Jacobs EJ, Chanock SJ, Fuchs CS, et al. Family history of cancer and risk of pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan). Int J Cancer 2010;127:1421-8. [Crossref] [PubMed]
- Austin MA, Kuo E, Van Den Eeden SK, et al. Family history of diabetes and pancreatic cancer as risk factors for pancreatic cancer: the PACIFIC study. Cancer Epidemiol Biomarkers Prev 2013;22:1913-7. [Crossref] [PubMed]
- Hemminki K, Li X. Familial and second primary pancreatic cancers: a nationwide epidemiologic study from Sweden. Int J Cancer 2003;103:525-30. [Crossref] [PubMed]
- Klein AP, Brune KA, Petersen GM, et al. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res 2004;64:2634-8. [Crossref] [PubMed]
- Dat NM, Sontag SJ. Pancreatic carcinoma in brothers. Ann Intern Med 1982;97:282. [Crossref] [PubMed]
- Ghadirian P, Simard A, Baillargeon J. Cancer of the pancreas in two brothers and one sister. Int J Pancreatol 1987;2:383-91. [PubMed]
- MacDermott RP, Kramer P. Adenocarcinoma of the pancreas in four siblings. Gastroenterology 1973;65:137-9. [PubMed]
- Friedman JM, Fialkow PJ. Familial carcinoma of the pancreas. Clin Genet 1976;9:463-9. [Crossref] [PubMed]
- Ehrenthal D, Haeger L, Griffin T, et al. Familial pancreatic adenocarcinoma in three generations. A case report and a review of the literature. Cancer 1987;59:1661-4. [Crossref] [PubMed]
- Klein AP, Beaty TH, Bailey-Wilson JE, et al. Evidence for a major gene influencing risk of pancreatic cancer. Genet Epidemiol 2002;23:133-49. [Crossref] [PubMed]
- Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000;343:78-85. [Crossref] [PubMed]
- Brune KA, Lau B, Palmisano E, et al. Importance of age of onset in pancreatic cancer kindreds. J Natl Cancer Inst 2010;102:119-26. [Crossref] [PubMed]
- Lorenzo Bermejo J, Hemminki K. Familial association of histology specific breast cancers with cancers at other sites. Int J Cancer 2004;109:430-5. [Crossref] [PubMed]
- Hiripi E, Bermejo JL, Li X, et al. Familial association of pancreatic cancer with other malignancies in Swedish families. Br J Cancer 2009;101:1792. [Crossref] [PubMed]
- Cote ML, Schenk M, Schwartz AG, et al. Risk of other cancers in individuals with a family history of pancreas cancer. J Gastrointest Cancer 2007;38:119-26. [Crossref] [PubMed]
- Hemminki K, Li X. Familial liver and gall bladder cancer: a nationwide epidemiological study from Sweden. Gut 2003;52:592-6. [Crossref] [PubMed]
- Wang L, Brune KA, Visvanathan K, et al. Elevated Cancer Mortality in the Relatives of Patients with Pancreatic Cancer. Cancer Epidemiol Biomarkers Prev 2009;18:2829-34. [Crossref] [PubMed]
- Thompson D, Easton DF. Breast Cancer Linkage Consortium. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst 2002;94:1358-65. [Crossref] [PubMed]
- Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 1999;91:1310-6. [Crossref] [PubMed]
- van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet 2005;42:711-9. [Crossref] [PubMed]
- Risch HA, McLaughlin JR, Cole DEC, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 2006;98:1694-706. [Crossref] [PubMed]
- Goggins M, Schutte M, Lu J, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res 1996;56:5360-4. [PubMed]
- Holter S, Borgida A, Dodd A, et al. Germline BRCA Mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol 2015;33:3124-9. [Crossref] [PubMed]
- Salo-Mullen EE, O’Reilly EM, Kelsen DP, et al. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer 2015;121:4382-8. [Crossref] [PubMed]
- Hu C, Hart SN, Bamlet WR, et al. Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol Biomarkers Prev 2016;25:207-11. [Crossref] [PubMed]
- Shindo K, Yu J, Suenaga M, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol 2017;35:3382-90. [Crossref] [PubMed]
- Hahn SA, Greenhalf B, Ellis I, et al. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst 2003;95:214-21. [Crossref] [PubMed]
- Couch FJ, Johnson MR, Rabe KG, et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev 2007;16:342-6. [Crossref] [PubMed]
- Slater EP, Langer P, Fendrich V, et al. Prevalence of BRCA2 and CDKN2a mutations in German familial pancreatic cancer families. Fam Cancer 2010;9:335-43. [Crossref] [PubMed]
- Zhen DB, Rabe KG, Gallinger S, et al. BRCA1, BRCA2, PALB2, and CDKN2A mutations in familial pancreatic cancer: a PACGENE study. Genet Med 2015;17:569-77. [Crossref] [PubMed]
- Catts ZA-K, Baig MK, Milewski B, et al. Statewide retrospective review of familial pancreatic cancer in delaware, and frequency of genetic mutations in pancreatic cancer kindreds. Ann Surg Oncol 2016;23:1729-35. [Crossref] [PubMed]
- Chaffee KG, Oberg AL, McWilliams RR, et al. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Murphy KM, Brune KA, Griffin C, et al. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: deleterious BRCA2 mutations in 17%. Cancer Res 2002;62:3789-93. [PubMed]
- Mocci E, Milne RL, Méndez-Villamil EY, et al. Risk of pancreatic cancer in breast cancer families from the breast cancer family registry. Cancer Epidemiol Biomarkers Prev 2013;22:803-11. [Crossref] [PubMed]
- Mersch J, Jackson MA, Park M, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2015;121:269-75. [Crossref] [PubMed]
- Streff H, Profato J, Ye Y, et al. Cancer incidence in first- and second-degree relatives of BRCA1 and BRCA2 Mutation Carriers. Oncologist 2016;21:869-74. [Crossref] [PubMed]
- Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet 2007;39:165-7. [Crossref] [PubMed]
- Jones S, Hruban RH, Kamiyama M, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 2009;324:217. [Crossref] [PubMed]
- Tischkowitz MD, Sabbaghian N, Hamel N, et al. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology 2009;137:1183-6. [Crossref] [PubMed]
- Slater EP, Langer P, Niemczyk E, et al. PALB2 mutations in European familial pancreatic cancer families. Clin Genet 2010;78:490-4. [Crossref] [PubMed]
- Hofstatter EW, Domchek SM, Miron A, et al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer 2011;10:225-31. [Crossref] [PubMed]
- Peterlongo P, Catucci I, Pasquini G, et al. PALB2 germline mutations in familial breast cancer cases with personal and family history of pancreatic cancer. Breast Cancer Res Treat 2011;126:825-8. [Crossref] [PubMed]
- Stadler ZK, Salo-Mullen E, Sabbaghian N, et al. Germline PALB2 mutation analysis in breast-pancreas cancer families. J Med Genet 2011;48:523-5. [Crossref] [PubMed]
- Ghiorzo P, Pensotti V, Fornarini G, et al. Contribution of germline mutations in the BRCA and PALB2 genes to pancreatic cancer in Italy. Fam Cancer 2012;11:41-7. [Crossref] [PubMed]
- Borecka M, Zemankova P, Vocka M, et al. Mutation analysis of the PALB2 gene in unselected pancreatic cancer patients in the Czech Republic. Cancer Genet 2016;209:199-204. [Crossref] [PubMed]
- Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov 2012;2:41-6. [Crossref] [PubMed]
- Grant RC, Selander I, Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology 2015;148:556-64. [Crossref] [PubMed]
- Takai E, Yachida S, Shimizu K, et al. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget 2016;7:74227-35. [PubMed]
- Roberts NJ, Norris AL, Petersen GM, et al. Whole genome sequencing defines the genetic heterogeneity of familial pancreatic cancer. Cancer Discov. 2016;6:166-75. [Crossref] [PubMed]
- Hemminki A, Tomlinson I, Markie D, et al. Localization of a susceptibility locus for Peutz-Jeghers syndrome to 19p using comparative genomic hybridization and targeted linkage analysis. Nat Genet 1997;15:87-90. [Crossref] [PubMed]
- Hemminki A, Markie D, Tomlinson I, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 1998;391:184-7. [Crossref] [PubMed]
- Jenne DE, Reimann H, Nezu J, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet 1998;18:38-43. [Crossref] [PubMed]
- van Lier MGF, Wagner A, Mathus-Vliegen EM, et al. High cancer risk in Peutz-Jeghers syndrome: a systematic review and surveillance recommendations. Am J Gastroenterol 2010;105:1258-64. [Crossref] [PubMed]
- Giardiello FM, Brensinger JD, Tersmette AC, et al. Very high risk of cancer in familial Peutz-Jeghers syndrome. Gastroenterology 2000;119:1447-53; author reply 1265. [Crossref] [PubMed]
- Korsse SE, Harinck F, van Lier MG, et al. Pancreatic cancer risk in Peutz-Jeghers syndrome patients: a large cohort study and implications for surveillance. J Med Genet 2013;50:59-64. [Crossref] [PubMed]
- Resta N, Pierannunzio D, Lenato GM, et al. Cancer risk associated with STK11/LKB1 germline mutations in Peutz-Jeghers syndrome patients: results of an Italian multicenter study. Dig Liver Dis 2013;45:606-11. [Crossref] [PubMed]
- Hearle N, Schumacher V, Menko FH, et al. Frequency and spectrum of cancers in the Peutz-Jeghers syndrome. Clin Cancer Res 2006;12:3209-15. [Crossref] [PubMed]
- Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med 1995;333:970-4. [Crossref] [PubMed]
- Ghiorzo P, Ciotti P, Mantelli M, et al. Characterization of ligurian melanoma families and risk of occurrence of other neoplasia. Int J Cancer 1999;83:441-8. [Crossref] [PubMed]
- Borg A, Sandberg T, Nilsson K, et al. High frequency of multiple melanomas and breast and pancreas carcinomas in CDKN2A mutation-positive melanoma families. J Natl Cancer Inst 2000;92:1260-6. [Crossref] [PubMed]
- Goldstein AM, Struewing JP, Fraser MC, et al. Prospective risk of cancer in CDKN2A germline mutation carriers. J Med Genet 2004;41:421-4. [Crossref] [PubMed]
- de Snoo FA, Bishop DT, Bergman W, et al. Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res 2008;14:7151-7. [Crossref] [PubMed]
- Potjer TP, Kranenburg HE, Bergman W, et al. Prospective risk of cancer and the influence of tobacco use in carriers of the p16-Leiden germline variant. Eur J Hum Genet 2015;23:711-4. [Crossref] [PubMed]
- Mukherjee B, Delancey JO, Raskin L, et al. Risk of non-melanoma cancers in first-degree relatives of CDKN2A mutation carriers. J Natl Cancer Inst 2012;104:953-6. [Crossref] [PubMed]
- Watson P, Lynch HT. Extracolonic cancer in hereditary nonpolyposis colorectal cancer. Cancer 1993;71:677-85. [Crossref] [PubMed]
- Aarnio M, Sankila R, Pukkala E, et al. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 1999;81:214-8. [Crossref] [PubMed]
- Pande M, Wei C, Chen J, et al. Cancer spectrum in DNA mismatch repair gene mutation carriers: results from a hospital based Lynch syndrome registry. Fam Cancer 2012;11:441-7. [Crossref] [PubMed]
- Samadder NJ, Smith KR, Wong J, et al. Cancer risk in families fulfilling the amsterdam criteria for lynch syndrome. JAMA Oncol 2017;3:1697-701. [Crossref] [PubMed]
- Geary J, Sasieni P, Houlston R, et al. Gene-related cancer spectrum in families with hereditary non-polyposis colorectal cancer (HNPCC). Fam Cancer 2008;7:163-72. [Crossref] [PubMed]
- Kastrinos F, Mukherjee B, Tayob N, et al. Risk of pancreatic cancer in families with Lynch syndrome. JAMA 2009;302:1790-5. [Crossref] [PubMed]
- Win AK, Young JP, Lindor NM, et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol 2012;30:958-64. [Crossref] [PubMed]
- Møller P, Seppälä TT, Bernstein I, et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome Database. Gut 2017. [Epub ahead of print]. [PubMed]
- Lowenfels AB, Maisonneuve P, DiMagno EP, et al. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J Natl Cancer Inst 1997;89:442-6. [Crossref] [PubMed]
- Howes N, Lerch MM, Greenhalf W, et al. Clinical and genetic characteristics of hereditary pancreatitis in Europe. Clin Gastroenterol Hepatol 2004;2:252-61. [Crossref] [PubMed]
- Rebours V, Boutron-Ruault MC, Schnee M, et al. Risk of pancreatic adenocarcinoma in patients with hereditary pancreatitis: a national exhaustive series. Am J Gastroenterol 2008;103:111-9. [Crossref] [PubMed]
- Raimondi S, Lowenfels AB, Morselli-Labate AM, et al. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract Res Clin Gastroenterol 2010;24:349-58. [Crossref] [PubMed]
- Masamune A, Kikuta K, Hamada S, et al. Nationwide survey of hereditary pancreatitis in Japan. J Gastroenterol 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Lowenfels AB, Maisonneuve P, Whitcomb DC, et al. Cigarette smoking as a risk factor for pancreatic cancer in patients with hereditary pancreatitis. JAMA 2001;286:169-70. [Crossref] [PubMed]
- van der Heijden MS, Brody JR, Gallmeier E, et al. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol 2004;165:651-7. [Crossref] [PubMed]
- van der Heijden MS, Brody JR, Dezentje DA, et al. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res 2005;11:7508-15. [Crossref] [PubMed]
- Villarroel MC, Rajeshkumar NV, Garrido-Laguna I, et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther 2011;10:3-8. [Crossref] [PubMed]
- Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005;434:913-7. [Crossref] [PubMed]
- McCabe N, Lord CJ, Tutt ANJ, et al. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol Ther 2005;4:934-6. [Crossref] [PubMed]
- Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 2010;376:235-44. [Crossref] [PubMed]
- Perkhofer L, Schmitt A, Romero Carrasco MC, et al. ATM deficiency generating genomic instability sensitizes pancreatic ductal adenocarcinoma cells to therapy-induced DNA Damage. Cancer Res 2017;77:5576-90. [Crossref] [PubMed]
- Jacob DA, Bahra M, Langrehr JM, et al. Combination therapy of poly (ADP-ribose) polymerase inhibitor 3-aminobenzamide and gemcitabine shows strong antitumor activity in pancreatic cancer cells. J Gastroenterol Hepatol 2007;22:738-48. [PubMed]
- Chen S, Wang G, Niu X, et al. Combination of AZD2281 (Olaparib) and GX15-070 (Obatoclax) results in synergistic antitumor activities in preclinical models of pancreatic cancer. Cancer Lett 2014;348:20-8. [Crossref] [PubMed]
- Porcelli L, Quatrale AE, Mantuano P, et al. Optimize radiochemotherapy in pancreatic cancer: PARP inhibitors a new therapeutic opportunity. Mol Oncol 2013;7:308-22. [Crossref] [PubMed]
- Pishvaian MJ, Wang H, Zhuang T, et al. A phase I/II study of ABT-888 in combination with 5-fluorouracil (5-FU) and oxaliplatin (Ox) in patients with metastatic pancreatic cancer (MPC). J Clin Oncol 2013;31:147. [Crossref]
- O’Reilly EM, Lowery MA, Segal MF, et al. Phase IB trial of cisplatin (C), gemcitabine (G), and veliparib (V) in patients with known or potential BRCA or PALB2-mutated pancreas adenocarcinoma (PC). J Clin Oncol 2014;32:4023. [PubMed]
- Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244-50. [Crossref] [PubMed]
- Olaparib in gBRCA Mutated Pancreatic Cancer Whose Disease Has Not Progressed on First Line Platinum-Based Chemotherapy - Full Text View - ClinicalTrials.gov [Internet]. Available online: https://clinicaltrials.gov/ct2/show/NCT02184195
- Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet 2009;41:986-90. [Crossref] [PubMed]
- Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 2010;42:224-8. [Crossref] [PubMed]
- Wolpin BM, Rizzato C, Kraft P, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet 2014;46:994-1000. [Crossref] [PubMed]
- Childs EJ, Mocci E, Campa D, et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat Genet 2015;47:911-6. [Crossref] [PubMed]
- Zhang M, Wang Z, Obazee O, et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget 2016;7:66328-43. [Crossref] [PubMed]
- Willis JA, Olson SH, Orlow I, et al. A replication study and genome-wide scan of single-nucleotide polymorphisms associated with pancreatic cancer risk and overall survival. Clin Cancer Res 2012;18:3942-51. [Crossref] [PubMed]
- Rizzato C, Campa D, Pezzilli R, et al. ABO blood groups and pancreatic cancer risk and survival: results from the PANcreatic Disease ReseArch (PANDoRA) consortium. Oncol Rep 2013;29:1637-44. [Crossref] [PubMed]
- Risch HA, Lu L, Wang J, et al. ABO blood group and risk of pancreatic cancer: a study in Shanghai and meta-analysis. Am J Epidemiol 2013;177:1326-37. [Crossref] [PubMed]
- Xu HL, Cheng JR, Zhang W, et al. Re-evaluation of ABO gene polymorphisms detected in a genome-wide association study and risk of pancreatic ductal adenocarcinoma in a Chinese population. Chin J Cancer 2014;33:68-73. [Crossref] [PubMed]
- Nakao M, Matsuo K, Hosono S, et al. ABO blood group alleles and the risk of pancreatic cancer in a Japanese population. Cancer Sci 2011;102:1076-80. [Crossref] [PubMed]
- Wolpin BM, Chan AT, Hartge P, et al. ABO blood group and the risk of pancreatic cancer. J Natl Cancer Inst 2009;101:424-31. [Crossref] [PubMed]
- Wolpin BM, Kraft P, Gross M, et al. Pancreatic cancer risk and ABO blood group alleles: results from the pancreatic cancer cohort consortium. Cancer Res 2010;70:1015-23. [Crossref] [PubMed]
- Mori A, Moser C, Lang SA, et al. Up-regulation of Krüppel-like factor 5 in pancreatic cancer is promoted by interleukin-1beta signaling and hypoxia-inducible factor-1alpha. Mol Cancer Res 2009;7:1390-8. [Crossref] [PubMed]
- Diaferia GR, Balestrieri C, Prosperini E, et al. Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 2016;35:595-617. [Crossref] [PubMed]
- Abjalimov IR, Zinovyeva MV, Nikolaev LG, et al. Expression of transcription factor genes in cell lines corresponding to different stages of pancreatic cancer progression. Dokl Biochem Biophys 2017;475:267-70. [Crossref] [PubMed]
- Kondratyeva LG, Chernov IP, Zinovyeva MV, et al. Expression of master regulatory genes of embryonic development in pancreatic tumors. Dokl Biochem Biophys 2017;475:250-2. [Crossref] [PubMed]
- Zhang D, Dai Y, Cai Y, et al. KLF2 is downregulated in pancreatic ductal adenocarcinoma and inhibits the growth and migration of cancer cells. Tumour Biol 2016;37:3425-31. [Crossref] [PubMed]
- Luo Z, Li Y, Zuo M, et al. Effect of NR5A2 inhibition on pancreatic cancer stem cell (CSC) properties and epithelial-mesenchymal transition (EMT) markers. Mol Carcinog 2017;56:1438-48. [Crossref] [PubMed]
- Flandez M, Cendrowski J, Cañamero M, et al. Nr5a2 heterozygosity sensitises to, and cooperates with, inflammation in KRasG12V-driven pancreatic tumourigenesis. Gut 2014;63:647-55. [Crossref] [PubMed]
- von Figura G, Morris JP 4th, Wright CV, et al. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut 2014;63:656-64. [Crossref] [PubMed]
- Campa D, Rizzato C, Stolzenberg-Solomon R, et al. TERT gene harbors multiple variants associated with pancreatic cancer susceptibility. Int J Cancer 2015;137:2175-83. [Crossref] [PubMed]
- Huang DS, Wang Z, He XJ, et al. Recurrent TERT promoter mutations identified in a large-scale study of multiple tumour types are associated with increased TERT expression and telomerase activation. Eur J Cancer 2015;51:969-76. [Crossref] [PubMed]
- Bojesen SE, Pooley KA, Johnatty SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013;45:371-84, 384e1-2.
- Hezel AF, Kimmelman AC, Stanger BZ, et al. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 2006;20:1218-49. [Crossref] [PubMed]
- van Heek NT, Meeker AK, Kern SE, et al. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am J Pathol 2002;161:1541-7. [Crossref] [PubMed]
- Bao Y, Prescott J, Yuan C, et al. Leucocyte telomere length, genetic variants at the TERT gene region and risk of pancreatic cancer. Gut 2017;66:1116-22. [Crossref] [PubMed]
- James MA, Wen W, Wang Y, et al. Functional Characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 Locus. PLoS One 2012;7:e36116. [Crossref] [PubMed]
- Ni Z, Tao K, Chen G, et al. CLPTM1L is overexpressed in lung cancer and associated with apoptosis. PLoS One 2012;7:e52598. [Crossref] [PubMed]
- Jia J, Bosley AD, Thompson A, et al. CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res 2014;74:2785-95. [Crossref] [PubMed]
- James MA, Vikis HG, Tate E, et al. CRR9/CLPTM1L regulates cell survival signaling and is required for ras transformation and lung tumorigenesis. Cancer Res 2014;74:1116-27. [Crossref] [PubMed]
- Adams JC, Seed B, Lawler J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J 1998;17:4964-74. [Crossref] [PubMed]
- Fernandez-Zapico ME, Lomberk GA, Tsuji S, et al. A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth. Biochem J 2011;435:529-37. [Crossref] [PubMed]
- Fan G, Sun L, Shan P, et al. Loss of KLF14 triggers centrosome amplification and tumorigenesis. Nat Commun 2015;6:8450. [Crossref] [PubMed]
- Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579-89. [Crossref] [PubMed]
- Wang J, Zhang J, Shen J, et al. Association of KCNQ1 and KLF14 polymorphisms and risk of type 2 diabetes mellitus: A global meta-analysis. Hum Immunol 2014;75:342-7. [Crossref] [PubMed]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707-13. [Crossref] [PubMed]
- Small KS, Hedman AK, Grundberg E, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet 2011;43:561-4. [Crossref] [PubMed]
- Barrett A, Pellet-Many C, Zachary IC, et al. p130Cas: a key signalling node in health and disease. Cell Signal 2013;25:766-77. [Crossref] [PubMed]
- Nikonova AS, Gaponova AV, Kudinov AE, et al. CAS proteins in health and disease: An update. IUBMB Life 2014;66:387-95. [Crossref] [PubMed]
- Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Dig Dis Sci 2007;52:1-17. [Crossref] [PubMed]
- Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet 2009;41:703-7. [Crossref] [PubMed]
- Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981-90. [Crossref] [PubMed]
- Harder MN, Ribel-Madsen R, Justesen JM, et al. Type 2 diabetes risk alleles near BCAR1 and in ANK1 associate with decreased β-cell function whereas risk alleles near ANKRD55 and GRB14 associate with decreased insulin sensitivity in the Danish Inter99 cohort. J Clin Endocrinol Metab 2013;98:E801-6. [Crossref] [PubMed]
- ’t Hart LM, Fritsche A, Nijpels G, et al. The CTRB1/2 locus affects diabetes susceptibility and treatment via the incretin pathway. Diabetes 2013;62:3275-81. [Crossref] [PubMed]
- Li D, Duell EJ, Yu K, et al. Pathway analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. Carcinogenesis 2012;33:1384-90. [Crossref] [PubMed]
- Offield MF, Jetton TL, Labosky PA, et al. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development 1996;122:983-95. [PubMed]
- Ahlgren U, Jonsson J, Jonsson L, et al. beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev 1998;12:1763-8. [Crossref] [PubMed]
- McKinnon CM, Docherty K. Pancreatic duodenal homeobox-1, PDX-1, a major regulator of beta cell identity and function. Diabetologia 2001;44:1203-14. [Crossref] [PubMed]
- Melloul D, Tsur A, Zangen D. Pancreatic duodenal homeobox (PDX-1) in health and disease. J Pediatr Endocrinol Metab 2002;15:1461-72. [Crossref] [PubMed]
- Hao HX, Xie Y, Zhang Y, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012;485:195-200. [Crossref] [PubMed]
- Magni M, Ruscica V, Buscemi G, et al. Chk2 and REGγ-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res 2014;42:13150-60. [Crossref] [PubMed]
- Nevanlinna H, Bartek J. The CHEK2 gene and inherited breast cancer susceptibility. Oncogene 2006;25:5912-9. [Crossref] [PubMed]
- Siołek M, Cybulski C, Gąsior-Perczak D, et al. CHEK2 mutations and the risk of papillary thyroid cancer. Int J Cancer 2015;137:548-52. [Crossref] [PubMed]
- Lawrenson K, Iversen ES, Tyrer J, et al. Common variants at the CHEK2 gene locus and risk of epithelial ovarian cancer. Carcinogenesis 2015;36:1341-53. [Crossref] [PubMed]
- Havranek O, Kleiblova P, Hojny J, et al. Association of Germline CHEK2 gene variants with risk and prognosis of non-hodgkin lymphoma. PloS One 2015;10:e0140819. [Crossref] [PubMed]
- Wang Y, Dai B, Ye D. CHEK2 mutation and risk of prostate cancer: a systematic review and meta-analysis. Int J Clin Exp Med 2015;8:15708-15. [PubMed]
- Lener MR, Kashyap A, Kluźniak W, et al. The Prevalence of Founder Mutations among Individuals from Families with Familial Pancreatic Cancer Syndrome. Cancer Res Treat 2017;49:430-6. [Crossref] [PubMed]
- Wu C, Miao X, Huang L, et al. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet 2011;44:62-6. [Crossref] [PubMed]
- Zheng J, Huang X, Tan W, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet 2016;48:747-57. [Crossref] [PubMed]
- Shi X, Ma C, Zhu Q, et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget 2016;7:25558-75. [Crossref] [PubMed]
- Ma C, Wu G, Zhu Q, et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget 2017;8:32696-705. [PubMed]
- Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer 2017;16:118. [Crossref] [PubMed]
- Yu J, Liu Y, Gong Z, et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget 2017;8:16621-32. [PubMed]
- Abdul-Rahman U, Győrffy B, Adams BD. linc00673 (ERRLR01) is a prognostic indicator of overall survival in breast cancer. Transcription 2017. [Epub ahead of print]. [Crossref] [PubMed]
- Sherman EA, Strauss KA, Tortorelli S, et al. Genetic mapping of glutaric aciduria, type 3, to chromosome 7 and identification of mutations in c7orf10. Am J Hum Genet 2008;83:604-9. [Crossref] [PubMed]
- Ito Y, Takeda T, Wakasa KI, et al. Expression of p73 and p63 proteins in pancreatic adenocarcinoma: p73 overexpression is inversely correlated with biological aggressiveness. Int J Mol Med 2001;8:67-71. [PubMed]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ 2011;18:1487-99. [Crossref] [PubMed]
- Danilov AV, Neupane D, Nagaraja AS, et al. DeltaNp63alpha-mediated induction of epidermal growth factor receptor promotes pancreatic cancer cell growth and chemoresistance. PloS One 2011;6:e26815. [Crossref] [PubMed]
- Flores ER, Sengupta S, Miller JB, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 2005;7:363-73. [Crossref] [PubMed]
- Kunisaki R, Ikawa S, Maeda T, et al. p51/p63, a novel p53 homologue, potentiates p53 activity and is a human cancer gene therapy candidate. J Gene Med 2006;8:1121-30. [Crossref] [PubMed]
- Park SL, Chang SC, Cai L, et al. Associations between variants of the 8q24 chromosome and nine smoking-related cancer sites. Cancer Epidemiol Biomarkers Prev 2008;17:3193-202. [Crossref] [PubMed]
- Goode EL, Chenevix-Trench G, Song H, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet 2010;42:874-9. [Crossref] [PubMed]
- Gudmundsson J, Sulem P, Gudbjartsson DF, et al. A common variant at 8q24.21 is associated with renal cell cancer. Nat Commun 2013;4:2776. [Crossref] [PubMed]
- Sur I, Tuupanen S, Whitington T, et al. Lessons from functional analysis of genome-wide association studies. Cancer Res 2013;73:4180-4. [Crossref] [PubMed]
- Tanskanen T, van den Berg L, Välimäki N, et al. Genome-wide association study and meta-analysis in Northern European populations replicate multiple colorectal cancer risk loci. Int J Cancer 2018;142:540-6. [Crossref] [PubMed]
- Hessmann E, Schneider G, Ellenrieder V, et al. MYC in pancreatic cancer: novel mechanistic insights and their translation into therapeutic strategies. Oncogene 2016;35:1609-18. [Crossref] [PubMed]
- Schleger C, Verbeke C, Hildenbrand R, et al. c-MYC activation in primary and metastatic ductal adenocarcinoma of the pancreas: incidence, mechanisms, and clinical significance. Mod Pathol 2002;15:462-9. [Crossref] [PubMed]
- Birnbaum DJ, Adélaïde J, Mamessier E, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer 2011;50:456-65. [Crossref] [PubMed]
- Lin WC, Rajbhandari N, Liu C, et al. Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res 2013;73:1821-30. [Crossref] [PubMed]
- Low SK, Kuchiba A, Zembutsu H, et al. Genome-wide association study of pancreatic cancer in Japanese population. PloS One 2010;5:e11824. [Crossref] [PubMed]
- Liang Y, Wu H, Lei R, et al. Transcriptional network analysis identifies BACH1 as a master regulator of breast cancer bone metastasis. J Biol Chem 2012;287:33533-44. [Crossref] [PubMed]
- Davudian S, Shajari N, Kazemi T, et al. BACH1 silencing by siRNA inhibits migration of HT-29 colon cancer cells through reduction of metastasis-related genes. Biomed Pharmacother 2016;84:191-8. [Crossref] [PubMed]
- Shajari N, Davudian S, Kazemi T, et al. Silencing of BACH1 inhibits invasion and migration of prostate cancer cells by altering metastasis-related gene expression. Artif Cells Nanomed Biotechnol 2017.1-10. [Crossref] [PubMed]
- Terris B, Blaveri E, Crnogorac-Jurcevic T, et al. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol 2002;160:1745-54. [Crossref] [PubMed]
- Yeh TS, Jan YY, Chiu CT, et al. Characterisation of oestrogen receptor, progesterone receptor, trefoil factor 1, and epidermal growth factor and its receptor in pancreatic cystic neoplasms and pancreatic ductal adenocarcinoma. Gut 2002;51:712-6. [Crossref] [PubMed]
- Prasad NB, Biankin AV, Fukushima N, et al. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res 2005;65:1619-26. [Crossref] [PubMed]
- Sunagawa M, Yamaguchi J, Kokuryo T, et al. Trefoil factor family 1 expression in the invasion front is a poor prognostic factor associated with lymph node metastasis in pancreatic cancer. Pancreatology 2017;17:782-7. [Crossref] [PubMed]
- Radiloff DR, Wakeman TP, Feng J, et al. Trefoil factor 1 acts to suppress senescence induced by oncogene activation during the cellular transformation process. Proc Natl Acad Sci U S A 2011;108:6591-6. [Crossref] [PubMed]
- Arumugam T, Brandt W, Ramachandran V, et al. Trefoil factor 1 stimulates both pancreatic cancer and stellate cells and increases metastasis. Pancreas 2011;40:815-22. [Crossref] [PubMed]
- Zhang Z, Chen Y, Tang J, et al. Frequent loss expression of dab2 and promotor hypermethylation in human cancers: a meta-analysis and systematic review. Pak J Med Sci 2014;30:432-7. [PubMed]
- Du L, Zhao Z, Ma X, et al. miR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene 2014;33:4307-15. [Crossref] [PubMed]
- Xie Y, Zhang Y, Jiang L, et al. Disabled homolog 2 is required for migration and invasion of prostate cancer cells. Front Med 2015;9:312-21. [Crossref] [PubMed]
- Cheong SM, Choi H, Hong BS, et al. Dab2 is pivotal for endothelial cell migration by mediating VEGF expression in cancer cells. Exp Cell Res 2012;318:550-7. [Crossref] [PubMed]
- Wang Z, Tseng CP, Pong RC, et al. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer. Characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J Biol Chem 2002;277:12622-31. [Crossref] [PubMed]
- Su Q, Wang Y, Zhao J, et al. Polymorphisms of PRLHR and HSPA12A and risk of gastric and colorectal cancer in the Chinese Han population. BMC Gastroenterol 2015;15:107. [Crossref] [PubMed]
- Park MY, Kim HS, Lee M, et al. FAM19A5, a brain-specific chemokine, inhibits RANKL-induced osteoclast formation through formyl peptide receptor 2. Sci Rep 2017;7:15575. [Crossref] [PubMed]
- Canto MI, Harinck F, Hruban RH, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 2013;62:339-47. [Crossref] [PubMed]


