Metallothionein 2A an interactive protein linking phosphorylated FADD to NF-κB pathway leads to colorectal cancer formation
Introduction
In the post-genome era, identification of disease related proteins by group or individual genes will provide a great opportunity for initial identification of the disease and then for the treatment with genetic markers by accurately (1). Tremendous increase incidence report of colorectal cancer has led to the search for genetic markers for early detection and then it may yield for better management. In spite of identification several proteins identified as genetic markers and including metallothionein (MT), have been identified as potential biomarkers in colorectal cancer (2). Human colorectal adenocarcinoma or colorectal cancer is one of the leading cancer and it is the third most commonly diagnosed cancer in the region and the world (3). Colorectal cancer is involved with many tumor suppressor genes and oncogenes which was recorded and directly involved in that particular cancer development (4). However, there is no single gene was directly involved in colorectal cancer has been found.
Detection of protein-protein interaction can be identified by the yeast two-hybrid system and it has been widely used by the researchers (5). It is well known fact that interaction partners are involved in any kind of biological function and it can be utilized for therapeutic purposes (6). Therefore, we taught to find out the biological roles of FADD by associated proteins and protein-protein interactions by a two yeast hybrid system known as GAL4-based yeast two-hybrid system. The FADD gene (GenBank accession number: FJ937782) was isolated mRNA and cloned the open reading frame into a plasmid in our laboratory (7,8). Using the mRNA reverse transcriptase technique, the full-length cDNA of the phosphorylated Fas associated death domain (pFADD) gene which is known as serine 194 in the C-terminal of FADD gene was generated. It contains 624 nucleotides, open reading frame that predicts 206 amino acids (9). Using the full length pFADD cDNA as bait to and for the interaction screened a human heart cDNA library. Finally there were six positive clones were identified by the interaction with the pFADD protein. In this study we report one protein which bound tightly and made a tremendous interaction, which encoded human MT2A.
MT was first isolated from the horse kidney in 1957 and it act as a group of proteins which binds to cadmium (Cd) (10-12). In mammals, MTs constitute a super family of non-enzymatic polypeptides, low molecular weight (6–7 kDa) with 61–68 amino acids. It is distinctive amino acid composition with low or no histidine, high cysteine content and no aromatic amino acid and a high content of sulfur and metals in the form of metal thiolate clusters (13-17). Four isoforms MT-I–MT-IV of mammalian MTs have been identified (10). It was recorded MT-II is the best characterized MT identified so far. It consist with 62 amino acids, and 30 percent of the residues are cysteines which will act as metal thiolate clusters in series of motifs: Cys-X-Cys, Cys-X-Cys-Cys, Cys-X-X-Cys (where X is any other amino acid than cysteine) (14,18,19). Over past decades it was recorded as MT-II have slowly been unraveled, it has multiple function such as broad range of functions, metal homeostasis, including cell cycle progression, ROS scavenging, regulation of Zn-containing proteins (e.g., p53), angiogenesis, immune defense responses and cell differentiation (14,15,20). In non-pathological tissues MT-II are mainly localized in the cytoplasmic proteins, and its localization varies with cell cycle progression which will react in the during the S and G2 phase (14,21). Further it was revealed that suggesting that altered levels high or low of MT-II could be expected to contribute to abnormal cell growth; and this will be an eye opening for therapeutic treatment. As seen in cancer, as well as in the acquisition of therapy resistance. So far the studies involvement with MT-II and prognostic marker is still not identified for response for treatment.
The interaction of pFADD with MT2A was demonstrated in vitro and in vivo co-immunoprecipitation (co-IP) experiments by glutathione S-transferase (GST)-pull-down assays and Hek293 cells respectively. Interaction of two proteins co-localized mostly to nuclei and observed slightly to cytoplasm, as shown by epifluorescence microscopy experiment. With this experiment can conclude that the function of MT2A may be more towards regulation of cell proliferation rather than apoptosis and over-expression of MT2A indicate that it can promote tumor genesis. On tumor necrosis factor (TNF) receptor 1 (TNFR1) activation, and IκBα gets phosphorylated and degraded, leading to p65/p50 nuclear translocation and transcriptional activation of nuclear factor-κB (NF-κB) target genes also involved in this interaction (22,23). Interaction was further confirmed by two proteins interaction on cell proliferation and apoptosis in colorectal cancer cells. It is already proved with the experiment that MT2A is an interaction partner for pFADD that correlates with the suppression of apoptosis and induction of cell proliferation.
Methods
Cloning and plasmid construction
The expression vectors for cyan fluorescence protein (CFP)-tagged MT2A or yellow fluorescence protein (YFP)-tagged FADD was constructed by inserting full-length MT2A or FADD in-frame with CFP or YFP into the HindIII and BamHI sites of pECFP-C1 and pEYFP-C1 vectors (Clontech), respectively. The pGBKT7 vectors which known as DNA binding domain (DNA-BD) of GAL4 and pGADT7 vector which is known as activation domain (AD) of GAL4, and the pGADT7-T, pGBKT7-Lam, pGBKT7-53 and pCL1 control plasmids were directly purchased from Clontech. Full length FADD gene of 208 amino acids was cloned into plasmid pGBKT7-FADD with respect to GAL4 DNA-BD, was created by PCR generated product cloned into EcoRI and the BamHI sites of pGBKT7. Similar manner pGADT7-FADD GAL4-AD construct was made with the PCR generated product of FADD into EcoRI and the BamHI sites of pGADT7. Library for screening with plasmid with GAL4-AD in the pACT vector which consisted with human heart cDNA library were purchased from Clontech. Plasmid pALEX-FADD; pALEX-death domain (pALEX-DD); pALEX-death effecter domain (pALEX-DED), pALEX-C terminal deletion of DD, and pALEX MT2A respectively used to express the recombinant protein GST-FADD, GST-DD, GST-DED, GST-C deletion and GST MT2A respectively with containing a GST tag, was constructed by subcloning the FADD and MT2A PCR-generated fragment into the EcoRI site and the SalI site of pALEX. MT2A protein-coding region was cloned for in vitro transcription and translation was cloned into pcDNA6-V5-HisB-HA. FADD and MT2A proteins to be expressed in human cell lines (Hek293 cells); gene coding region was cloned into pcDNA6-V5-HisB-HA and pcDNA6-V5-HisB-FLAG, respectively. Antisense FADD gene was constructed by inserting the coding region into plasmid pcDNA3.1(+) − FADD by PCR product into the EcoRI site and the BamHI site of pcDNA3.1(+).
Bacterials and yeast strains
Library screening for protein and protein interactions Matchmaker GAL4 two-hybrid system 3 was purchased from Clontech. Yeast strain AH109 which was purchased consisted with (MATa, trp1–901, leu2–3, 112, ura3–52, his3–200, gal4D, gal80D, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-LacZ). AH109 had special features with eliminates false positives by using three reporters ADE2 for nutrition selection, HIS3 reduces false positives and MEL1 (or LacZ) encodes α- and β-galactosidase can be identified directly on X-α-gal indicator plates. For expression and plasmid construction was mainly done by Escherichia coli strains DH5α and BL21 respectively.
Cell culture and transfection
Human colorectal adenocarcinoma (Colo 205) cells, HeLa cell line, and Hek293 cells were purchased from the American Type Culture Collection [(ATCC), Philadelphia, PA, USA]. Colo 205 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% (v/v) fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA). Hek293 cells were grown in Medium 200 (Cascade Biologics, Portland, OR, USA) supplemented with Low Serum Growth Supplement (LSGS). All cells were cultured in a humidified CO2 incubator at 37 °C. Exponentially growing cells were dispersed with trypsin, seeded at 2×105 cells/35 mm glass bottom dish in 1.5 mL of culture medium. The transfection of CFP and YFP fusion protein constructs were carried out using Lipofectamine 2000 (Gibco).
Yeast two-hybrid library screening
Plasmid pGBKT7-FADD was used as bait in two-hybrid screens of human heart cDNA libraries by the Matchmaker two-hybrid system 3 protocol (Clontech). The yeast strain AH109 was transformed with the pGBKT7-FADD by the lithium acetate method and expression of the bait was confirmed by western blotting. A human heart cDNA library in the pACT2 vector (Clontech) was transformed into the yeast strain expressing the bait protein. Transformants expressing both the bait and interacting prey proteins were selected on medium lacking tryptophan, leucine, histidine, and adenine (Sigma). Plates were incubated at 30 °C for 5–7 days and tested for β galactosidase activity using the filter lift assay. Approximately 3×106 colonies were screened and 17 positive clones were identified. The cDNA inserts of the positive clones were amplified by PCR using primers complementary to the pACT2 vector (5' T ACC ACT ACAATG GAT 3' and 5' GTG AAC TTG CGG GGT TTT TCA GTA TCT ACG A 3'). Subsequently, the pACT2-cDNA constructs were isolated from positive yeast colonies, as recommended by the supplier, transformed into super-competent E. coli DH5α by electroporation, grown under selection, re-isolated, and analyzed by restriction digests. The unique purified constructs were then re-tested against the original pGBKT7-FADD bait construct. To ensure that the interactions were specific, the positive clones were also tested against an irrelevant bait protein lamin C and grown on selective plates lacking tryptophan, leucine, histidine, and adenine to test the specificity of interactions. The positive inserts were sequenced and analyzed by comparison to the GenBank sequence data bank.
Preparation of recombinant proteins and in vitro transcription and translation
For the GST-FADD binding experiments, the E. coli strain BL21 was transformed with pALEX-FADD; pALEX-DD; pALEX-DED and pALEX-C terminal deletion of DD, respectively, and it was grown in LB medium containing ampicillin (100 μg/mL). After 2 hours of induction, by adding l mM isopropyl-β-D-thiogalactopyranoside (IPTG) at 37 °C in a 500-mL of culture of E. coli with GST fusion protein in BL21 culture was harvested. Harvested bacterial pellet re-suspended in 20 mL phosphate-buffered saline (PBS) containing 1% of Triton X-100 and 0.5 mM dithiothreitol later which was sonicated thoroughly (10×30 s). After mixing gently for half an hour at 4 °C, spun the lysate was at 12,000 rpm for 10 min at 4 °C. The induced GST fusion protein was purified by supernatant was bound to 250 μL prewashed glutathione-Sepharose beads (Amersham Pharmacia Biotech) for an hour at 4 °C, by this GST fused protein will bound to the beads. Later beads were washed with PBS and the quantity and purity of the GST-fusion protein were analyzed by SDS-PAGE. In vitro translation reaction of pcDNA6-V5-HisB-FLAG-MT2A was carried out using the TNT Quick Coupled Transcription/Translation Systems (Promega) according to the manufacturer’s instructions (24).
GST-pull-down assay
Glutathione-Sepharose beads were incubated with GST-FADD or GST alone for binding in vitro translated pcDNA6-V5-HisB-FLAG-MT2A in a binding buffer on ice for 2 hours period (100 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 0.1% NP-40). Bounded beads were separated by centrifugation and washed several times mostly 4 times with the fresh buffer. This was done at the room temperature, and the product was re-suspended in 20 μL SDS sample buffer, and boiled at 100 °C for 3 min for run the electrophoresis. The protein separation was done on a 12% gel by SDS-PAGE electrophoresis and it was detected by western blotting using anti-FLAG monoclonal antibody (Sigma-Aldrich), peroxidase-conjugated goat anti-mouse IgG secondary antibody, and the western blotting luminal reagent (Santa Cruz Biotechnology) (24).
co-IP
Immunoprecipitation experiments were done in the Hek293 cell lines and plasmids were co-transfected with pcDNA6-V5-HisB-FLAGMT2A (10 μg) and pcDNA6-V5-HisB-HA-FADD (3.3 μg) with or without the control plasmids which is known as pcDNA6-V5-HisB-HA and pcDNA6-V5-HisBFLAG, it was done in 100-mm dishes using Lipofectamine 2000 (Invitrogen). After 48 hours of transfection, cells were removed from the plates by solubilized with 1 mL of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet p40, and 0.5% sodium deoxycholate) (Roche) on ice for 30 min. Other than the proteins, this is known as insoluble materials were removed by centrifugation for 20 s at 12,000 rpm at 4 °C. Proteins which were in the supernatants were collected and measured its concentration using the Bradford method and adjusted to a final concentration of 1 mg/mL. For co-IP supernatants were incubated or precleared with protein G (Roche) for 3 h at 4 °C. Out of the product 500 μL lysates were then incubated with 3 μg anti-HA (Roche) or 6 ìg anti-FLAG (Sigma) antibodies coupled to sepharose beads overnight at 4 °C on a rocking platform. The incubated product samples were subjected to be washed with wash buffer (wash 1: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet p40, and 0.5% sodium deoxycholate; wash 2: 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.1% Nonidet p40, and 0.05% sodium deoxycholate; wash 3: 10 mM Tris-HCl, pH 7.5, 0.1% Nonidet p40, and 0.05% sodium deoxycholate). To find out the interactions proteins retained on the beads were eluted by SDS sample buffer and subjected to electrophoresis by SDS-PAGE on a 12% gel, followed by immunoblot analysis with an anti-FLAG monoclonal antibody or the anti-HA monoclonal antibody (24).
Western blot analysis
The cells were solubilized with 0.5 mL of lysis buffer (Roche Molecular Biochemicals, Mannheim, Germany) on ice for 30 min. The insoluble fraction was removed by centrifugation for 10 min at 12,000 rpm at 4 °C. The supernatants were collected and supplemented with cOmplete Mini Protease Inhibitor Cocktail (Roche Molecular Biochemicals, Mannheim, Germany), and their protein concentrations were measured using Bradford method and 50 μg was used for western blotting. Protein extracts were separated by 8% SDS-PAGE and then electrophoretically transferred to nitrocellulose membranes (Hybond C, Amersham, Piscataway, NJ, USA). Membranes were blocked with 5% nonfat milk for 1 h and then incubated with anti-human FADD monoclonal antibody (0.2 µg/mL; Biodesign, Maine, USA) or anti-HA monoclonal antibody (0.5 µg/mL; Santa Cruz, California, USA), or anti-MT2A antibody (0.2 µg/mL; Sigma, St. Louis, MO, USA) for 1 h at room temperature. The membranes were washed 3 times with Phosphate Buffered Saline Tween-20 (PBST) followed by incubation for 1 h with HRP conjugated secondary antibodies. For detection of other proteins anti-goat horseradish antibody (0.1 µg/mL; Maixin, Fuzhou, China) was used as secondary antibody. The membrane was washed and then developed with enhanced chemiluminescence reagent (Amersham Life Science, USA) and exposed with Kodak X-Omat Blue film (NEN Life Science Products, Boston, MA, USA).
Detection of apoptosis
The cell lines (~1×105) treatment with over-expression of pRK5 (control), MT2A, pFADD, and MT2A-pFADD were washed twice with cold PBS and resuspended in 100-mL binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4). Annexin V-FITC (0.1 µg) was added according to the manufacturer’s instruction (Caltag Laboratories, Burlingame, CA, USA), and the cells were incubated in the dark for 10 min. Propidium iodide (10 µL, 50 µg/mL) was added and the cells were incubated for 15 min at room temperature. Finally, 400 mL of binding buffer was added to each tube, and the stained cells were analyzed by flow cytometry (FACSCalibur, Becton Dickinson; Denderstraat, Belgium) with CellQuest software within 1 h. Ten-thousand cells were recorded per assay.
Transient transfection and luciferase assays
The MMP-9 promoter, MMP-9 mAP-1, and MMP-9 mNF-κB constructs cloned into the pGL3-Basic luciferase vector (Promega) were purchased from Promega. Hek293 cells were transiently co-transfected with pCMV-β-gal and pGL3-MMP-9-Luc or pGL3-MMP-9 mAP-1-Luc or pGL3-MMP-9 mNF-κB-Luc, using the Lipofectamine 2000 reagent (Invitrogen, San Diego, CA, USA) according to the manufacturer’s protocol. Cells were then lysed, and luciferase activity was measured using a luminometer (Luminoskan Ascent; Thermo Electron Co., Waltham, MA, USA). Luciferase activity was normalized to β-galactosidase activity in cell lysates and was expressed as an average of three independent experiments (25).
Nuclear extractions and electrophoretic mobility shift assay (EMSA)
Nuclear extracts were prepared as previously described (26). Shortly, cells were washed, scraped, and collected by centrifugation at 2,500 rpm for 5 min at 4 °C. Cells were resuspended in low salt buffer, lysed for 15 min on ice, followed by addition of a 10% Igepal CA-630 solution and centrifugation. The pelleted nuclei were resuspended in high salt buffer and nuclear supernatants were obtained by centrifugation. DNA-binding activity of NF-κB was analyzed by EMSA and the following sequence was used as specific oligomer for NF-κB: 5'-AGTTGAGGGGACTTTCCCAGGC-3' (sense). Single-stranded oligonucleotides were labeled with γ-[32P]-ATP by T4-polynucleotide kinase (MBI Fermentas 17 GmbH, St. Leon-Rot, Germany), annealed to the complementary oligomer strand and purified on Sephadex columns (Illustra Nick Columns, GE Healthcare, Piscataway, NJ, USA). Binding reactions containing 5 µg nuclear extract, 1 µg Poly(dI:dC) (Sigma), labeled oligonucleotide (10,000 cpm) and 5× binding buffer were incubated for 30 min on ice. Binding complexes were resolved by electrophoresis in non-denaturing 6% polyacrylamide gels using 0.5× TBE as running buffer and assessed by autoradiography. For EMSA super shift experiments, binding reactions containing 2.5 µg nuclear extract were incubated for 30 min at room temperature with 4 µg of the following antibodies before addition of labeled oligonucleotides: anti-p65 (sc-372X from Santa Cruz) (27,28).
Sensitized emission FRET measurement with three-channel microscopy
HeLa cells were plated onto 0.17 mm thick bottom glass dishes and were transiently transfected with Lipofectamine 2000 (Gibco) 24 hours later. The cells were washed twice with PBS (pH 7.4) and covered with 1 mL fresh medium. Images were then taken with an Olympus IX81 inverted microscopy equipped with a 60×, NA =1.45 oil immersion objective lens and cooled-coupled device. Excitation light was delivered by an X-cite light source. For imaging, the image-pro plus software version 6.0 (Media Cybernetics, Rockville, MD, USA) was used. In most experiments, the excitation intensity was attenuated down to 25% of the maximum power of the light source. Images were acquired using the 1×1 binning mode and 400 ms integration times. For quantitative FRET measurements, the method of sensitized FRET was performed as described in detail earlier (29-31). Images were acquired sequentially through YFP, FRET and CFP filter channels. Here, the filter sets used were YFP (S500/20 nm; Q515lp; S535/30 nm, Chroma), CFP (S436/20 nm; Q455lp; S480/40 nm, Chroma) and FRET (S436/20 nm; Q455lp; S535/30 nm, Chroma). The background images were subtracted from the raw images before carrying out FRET calculation. Corrected FRET (FRETC) was calculated on a pixel-by-pixel basis for the entire image using the following equation: FRETC = FRET − (a × YFP) − (b × CFP), where FRET, CFP and YFP correspond to background-subtracted images of cells co-expressing CFP and YFP acquired through the FRET, CFP and YFP channels respectively. The “a” and “b” are the fractions of bleed-through of YFP and CFP fluorescence through the FRET filter channel, respectively. In our system, a=0.16±0.02, b=0.22±0.01. We used the following equation: FRET ratio (FR) = [FRET − (b × CFP)]/(a × YFP), a relative value that varies with changes in energy transfer to quantify the FRET signal. FR represents the fractional increase in YFP emission due to FRET. Thus, in the absence of energy transfer, FR has a predicted value of 1 (27,32).
Cell proliferation assay
The effects of pFADD + MT2A and FADD + MT2A on Hek293 cell proliferation were assessed by the MTT assay. In the exponential growth phase were seeded into a 96-well plate at a density of 5,000 cells per well. After 24 h, pFADD or FADD was added to a final concentration of 1, 5, or 10 µg/mL respectively. The cells were incubated at 37 °C for 48 h, then the cell viability was determined by the colorimetric MTT [3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazolium bromide] assay at wavelength 570 nm by TECAN Safire Fluorescence Absorbance and Luminescence Reader (Vienna, VA, USA). The cell viability was calculated according to the formula: cell viability (%) = average A570 nm of treated group/average A570 nm of control group ×100%.
Ethics statements
Six-week-old female athymic nude mice, which were purchased from the Vitalriver Animal Center (Vitalriver, Beijing, China), were housed in environmentally controlled conditions (22 °C, a 12-h light/dark cycle with the light cycle from 6:00 to 18:00 and the dark cycle from 18:00 to 6:00) and maintained on standard laboratory chow. Animal welfare and treatment were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals (the Ministry of Science and Technology of China, 2006) and all experimental protocols were approved under animal protocol number SYXK(Su)2009-0017 by the Animal Care and Use Committee of College of Life Sciences, Nanjing University.
RNA interference
Small interfering RNA (siRNA) was synthesized by Bosai (Beijing, China) with the following sequences: MT-2A, and pFADD with the 5'-CCG GCT CCT GCA AAT GCA AAG AGT G-3' and 5'-GAT CCC GTC ACA TAT GAC GTT AAA-3' respectively. Cells, grown to 50% confluence were transfected using oligofectamine transfection reagent (Invitrogen, Carlsbad, CA, USA) with MT-2A or scrambled siRNA according to the manufacturer’s instructions.
In vivo animal tumor model experiment
Female athymic nude mice (age 6 weeks) were obtained from the Vitalriver Animal Center and were acclimatized to local conditions for 1 week. Logarithmically growing human Colo 205 cells were harvested by trypsinization and suspended in PBS at a density of 1×107 cells/mL. Then, 100 mL of the single-cell suspension were injected subcutaneously into the right dorsum of nude mice. All tumor bearing mice were divided randomly into groups of 8–10, and treatment was initiated on day 10 when the volume of tumor reached about 40–50 mm3. The mice were injected intraperitoneally (i.p.) with control siRNA, MT RNAi, and pFADD RNAi day 12 and day 20, daily for 3 days. Tumor measurements were converted to tumor volume (V) as follows: L × W2 ×0.52, where L and W are the length and width, respectively. Measurements were taken by the vernier caliper. All procedures followed approval of the Institutional Animal Care Committee.
Statistics
All results in this study are expressed as means ± SE values. Student’s test was used to find the significance of differences among the means of various groups. Differences which reflect the significance difference were considered at P<0.05.
Results
A yeast two-hybrid screen of cellular proteins interacting with the pFADD protein
To detect the proteins which interact with the FADD protein, a fusion protein construct of the GAL4 DNA-BD with the FADD (open frame 1–624 bp) was cloned and interact as bait for the screening of a human heart tissue yeast two-hybrid cDNA library. Out of the 3×106 transformants screened 92 clones which were grown in the lack of tryptophan, leucine, histidine, and adenine and finally β-galactosidase activity was expressed. After the protein-protein interaction pACT2/cDNA plasmids were extracted and re-tested for the specificity of β-galactosidase expression assay. After re-transformation, with the outcome of protein interaction six independent positive clones were identified and sequenced (Table 1). The genes which was identified by the protein interaction assay known as yeast two-hybrid approach were metallothionein 2A (MT2A), humaninin 1, collagen type I alpha 1, cardiac myosin BP-C, HS chromosome 16 and mitochondrial binding protein. In this paper we focus on the characterization of the association of FADD with MT2A.

Full table
pFADD and MT2A interact in vitro
The GST-pFADD; GST DD, GST DED fusion protein were expressed in the bacteria strain E. coli BL21 and protein were purified by glutathione-Sepharose beads (Figure 1A,B). To assess and reflect whether the yeast protein-protein interaction data reflected direct binding between FADD and MT2A or not, the further study on ability of these proteins to interact in vitro was tested. In vitro translated MT2A was confirmed tested for interaction outside the cell binding with glutathione-Sepharose beads either coupled with GST alone or GST-FADD. After external interaction following with intensive washing, bound proteins to beads were eluted and analyzed by run on electrophoresis with SDS-PAGE, and confirmed by western blot. As shown in the given Figure 1C, MT2A was tightly bound to GST-pFADD but not to the control used as GST protein (Figure 1C).
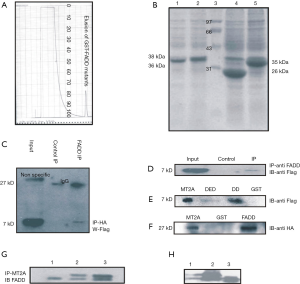
The above results are consistent with the yeast results and further confirmed the interaction between pFADD and MT2A. Aliquots of in vitro-translated pFADD were introduced with glutathione-Sepharose beads containing bound GST-pFADD mutants (DD/DED) or GST to find out the domain interaction. Input (MT2A) was the positive control. GST alone was used as a negative control, and detected by the anti-FLAG antibody. MT2A was observed to interact with FADD death domain as shown in the Figure 1D,E,F. In mutant immuno precipitates DD interact with MT2A was confirmed by GST pull down assay.
pFADD interacts with MT2A in vitro
Furthermore to confirm the interaction we investigate whether FADD and pFADD and MT2A occurred in vitro. This experiment was performed by well-known experiment co-IP of the complex from Hek293 cells by expression of the two proteins. HA epitope tagged FADD was expressed and transiently co-expressed in Hek293 cells together with pcDNA6-V5-HisB-FLAG-MT2A and the cell lysate with a high protein expression of HA-FADD, HA-DD; HA-DED. These results are consistent with the interaction between FADD and MT2A (Figure 1E,F). Aliquots of in vitro-translated pFADD were introduced with protein A mutants (DD/DED) or domain interaction. Input (MT2A) was the positive control. GST alone was used as a negative control, and detected by the anti-FLAG antibody. MT2A was observed to interact with FADD death domain as shown in the Figure 1F centre image and the bottom image. In mutant immuno precipitates DD interact with MT2A (Figure 1F). Co-IP of FADD and MT2A the MT 2A interaction in HeLa cells was confirmed by endogenous co-IP (Figure 1G), Furthermore, co-IP of pFADD and MT2A the MT 2A interaction in HeLa cells was confirmed by over expression co-IP (Figure 1H).
FRET measurements of FADD and MT2A in living cells with three-channel fluorescence microscopy
In order to analyze pFADD and MT2A self-association in vivo, sensitized emission FRET detection method which was based on three-channel fluorescence microscopy was used. CFP and YFP were inserted into the N-terminal of full length MT2A and FADD respectively. (See “Methods”) spectral profiles of CFP-tagged MT2A and YFP-tagged FADD were mostly identical to the reported spectra for the two fluorescence proteins. In addition, functional studies show that the biological functions of CFP-MT2A or YFP-FADD are not changed compared with MT2A and FADD (data not shown). To measure the steady-state FRET, cells transfected with different fusion proteins were imaged using an inverted epifluorescence microscopy through CFP, YFP, and FRET filter channels. The FRET efficiency was quantified as the FR (see “Methods”). To ensure that our recording system could reliably detect FRET, we first carried out some control experiments. As shown in Figure 2A, cells co-expressing CFP and YFP, which served as a negative control, showed no FRET with FR =1.03±0.02 (n=78) (Figure 2B). On the other hands, cells co-expressing the CFP-YFP concatemer, a positive control for FRET, showed significant increase in FR (FR =5.18±0.02, n=90) value (Figure 2B). At the same time strong FRET signal could be detected both in the cytoplasm and nuclear of cells. Compared with negative control, cells co-expressing CFP-MT2A and YFP-FADD also showed significant FRET signal both in the cytoplasm and nuclear of cells with FR =1.27±0.02 (n=50) (Figure 2B).

Characterization of the biological activity of pFADD, FADD with MT2A
In order to assess the effect of pFADD + MT2A on cell proliferation in vitro, endothelial cell proliferation assay was performed. As shown in the MTT assay Figure 2C, both FADD + MT2A displayed a dose-dependent inhibitory effect on cell pro proliferation, and pFADD + MT2A showed a more potent inhibitory effect than FADD + MT2A (P<0.05). The concentration of FADD + MT2A was about ED50 was approximately 10 µg/mL.
FADD and phosphorylation of FADD activates the NF-κB pathway
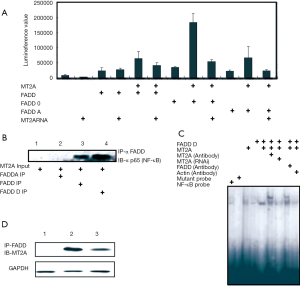
Next, we aimed at identifying the underlying molecular mechanisms responsible for the pFADD and MT 2A stimulated cell proliferation. To this end, we examined the effect of MT2A and FADD on NF-κB signaling, because pFADD (FADD-D)-mediated down regulation of MT2A proteins has been reported to lead to NF-κB activation from this study. To monitor activation of the canonical NF-κB pathway, we analyzed FADD phosphorylation. FADD, FADD-D and FADD-A (Ser-to-Ala substitution at position 194 to mimic nonphosphorylated FADD), were used in this assay and over expression of pFADD with MT2A stimulation accompanied by a slight increase in NF-κB protein levels (Figure 3A,B) when it was done in co-IP, IP FADD and IB with anti p65 which represent NF-κB. To analyze that NF-κB subunits translocate into the nucleus, we prepared cytosolic and nuclear extracts. FADD, pFADD stimulated with MT2A nuclear translocation and it was primarily triggered p65 translocation (Figure 3B). DNA-binding assays showed that FADD and MT2A stimulated NF-κB DNA binding within the first hours of stimulation and over a prolonged time up to at least 24 h (data not shown). Interestingly, EMSA super shift assay revealed that NF-κB DNA-binding complexes on stimulation with FADD, FADD-D with MT were mainly due to a new protein-protein interaction happens antibodies to nuclear extracts resulted in a super shift (for p65 antibody) of DNA-binding complexes (Figure 3C). To determine whether NF-κB DNA-binding leads to transcriptional activation of NF-κB target genes we performed luciferase assay. Phosphorylated FADD (FADD-D) and MT2A significantly increased NF-κB transcriptional activity. It was further confirmed with MT2A knockdown assay decreases the Luciferase assay. These experiments show that phosphorylated FADD activates NF-κB signaling and in particular the non-canonical pathway in HeLa cells resulting in enhanced NF-κB transcriptional activity. Finally we checked the endogenous interaction between pFADD and MT2A which the Figure 3D indicates that it was interact without induction of proteins.
Effect of FADD gene and MT2A gene on cell proliferation of HeLa cells
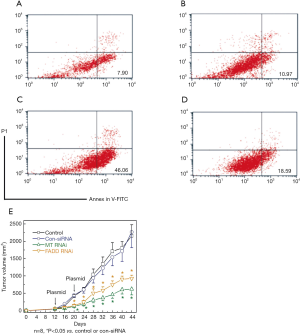
In initial experiments, the ability of HeLa cells to express the FADD and MT2A in experimental conditions was examined. We selected HeLa cell line to be transfected with pRK5(+) − FADD (Figure 4A); pRK5(+) − MT2A (Figure 4B) or co-transfected with pRK5(+) − MT2A (Figure 4C) with pRK5(+) FADD (Figure 4D) and the latter group was then transfected with pRK5(+) vector were used as control. After 24 h, apoptosis cells were characterized using Annexin V/propidium iodide staining. FADD alone could induce significant apoptosis in a dose-dependent manner, the apoptosis rate was (46.06%) at a dose of 10 μg/mL and reached the plateau. FADD inhibited proliferation and lead towards apoptosis and MT2A promoted cell proliferation (P<0.05). Cell apoptosis was inhibited in the group of MT2A co-transfected with pFADD compared with control groups and cell proliferation was significantly enhanced (P<0.05) (Figure 4D). Furthermore, FADD reduced the promotion of cell proliferation, and pFADD with MT2A these two proteins were positively associated on cell proliferation. In order to assess the effect of pFADD and FADD on MT2A in vitro, Hek293 cell proliferation assay was performed. As shown in the MTT assay (Figure 2A), both pFADD + MT2A and FADD + MT2A displayed a dose-dependent inhibitory effect on cell proliferation, and pFADD on MT2A showed a more potent inhibitory effect on apoptosis (P<0.05).
Inhibition of tumor growth
To determine whether knockdown of MT2A and pFADD could improve the antitumor activity, we used human colorectal adenocarcinoma cell line, Colo 205, in athymic nude mice (Figure 4E). From day 12 and 20 on, the mice were injected i.p. daily for 3 days with MT2A RNAi, FADD RNAi, control plasmid siRNA at 1.25 mg/kg/day dose. Both MT RNAi and FADD RNAi significantly inhibited the growth of Colo 205 solid tumors (on days 20–44, P<0.05, compared with control). At the same dose MT RNAi resulted in more significant inhibition of tumor growth than wild-type of Colo 205 (P<0.05, compared with wild-type Colo 205 at 1.25 mg/kg/day dose), while the antitumor activity of MT RNAi and pFADD RNAi was similar.
Conclusions
Increase incidence of death was recorded, due to metastasis is the cause for colorectal cancer, therefore strategies level of involvement for the development of treatment for the said cancer is needed or consider as the great importance. MTs have been identified as the main implicated in colorectal cancer study of this nature and progression as oncogenic proteins, which leads to promoting cell proliferation in several types of cancers (33). One of the greatest achievement in this century was the human genome project, which providing a wealth of information about genes that are related to disease or not which comprise with sequences of individual genes, reached completion (34,35), the track of research was diverted the focus of research into identifying the structure, function, and interactions of the proteins which linked to human diseases (36-38).The pFADD was a novel candidate cell proliferation gene which was associated with MT2A in colorectal cancer which was cloned to identify the interaction in our laboratory. Identifying the molecular level function of pFADD gene may provide golden opportunities to window of frame to elucidate the colorectal cancer mechanisms and its role in tumor development and progression. To answer the key question about a protein, in cellular lever to identify when and where it is expressed, and finally to identify the main roles of its with which other proteins it interacts (39). In this study we searched for associated proteins with a yeast two-hybrid system using the FADD cDNA fragment as bait. On screening a human heart cDNA library, we identified six putative clones as associated proteins including MT2A, humaninin 1, collagen type I alpha 1, cardiac myosin BP-C, HS chromosome 16 and mitochondrial binding protein since human MTs are closely linked with cancer (39). Furthermore it is possible by doing this research for us to understand the cellular level how it functions of the FADD protein together with binding to MT2A. Protein interaction with the yeast two-hybrid system provides sometime false interaction and therefore considered as only potential interactions will yield, therefore we need to be confirmed by further biological experimentation (40).Therefore, we performed GST-pull-down assays in vitro, co-IP experiments as inside the cell, and co-localization of the two proteins in vivo using double immunofluorescence staining to test the association of pFADD with MT2A. Finally based on what we got our results both in vivo and in vitro experiments shows and supported the interaction between pFADD and MT2A.
MTs are well known as a protein with a group of low molecular weight, rich with cysteine in the protein, as well as metal ion-binding proteins with the promotion of enhanced cell proliferation in colorectum carcinoma of the colon. Since MTs main role in oncogenesis and tumor progression is poorly studied and therefore therapy responses and patient prognosis is not clearly demonstrated, but it can protect against DNA damage, oxidative stress and increase proliferation studies were done intensively. Furthermore increased expression of MT-11 mRNA and protein studies shows that it is involved in various tumors such as ovary, urinary bladder, cervical, lung, and pancreatic cancers so on. Some studies showed that MT-11 expression correlates with tumor grade/stage. Although the down regulation of MT-2A by siRNA in Colo 205 cells results in the induction of growth and arrest apoptosis, the exact mechanism by which MT-2A affects cancer cell invasion in Colo 205 cancer module has been well elucidated in this study.
MT-11 highly expressed in tumor cells in cellular level in cytoplasm and/or nuclei in colorectal malignancies has been observed in both the tissue biopsies in patient with or without irradiation (41,42), further reported surrounding cells produced very low amount of express any detectable MT-II protein (41). It has been observed that MT-11 protein expression never changed in the cancer cells with radiation treatment (42). In addition, a significant decrease in the amount of MT-II protein in adenomas and carcinomas, as compared with normal (healthy) colorectal mucosa, has been reported (21,43). Above studies reflect that there is no correlation between protein expression of MT-11 with tumor stage, grade, or patient survival (41), and MT thus appears to have no impact on prognosis in colorectal cancer. But in our study it was revealed that MT-II is highly expressed in Colo 205 animal model.
In this study showed that transfection of pFADD induced cell proliferation in Hek293 cells and inhibited cell apoptosis. Observation was made that group of Hek293 cells transfected with FADD gene alone indicated high apoptotic rate and transfected with the MT2A gene alone there were high amount of cell numbers and promoted cell proliferation. In the group of Hek293 cells cotransfected with pFADD and MT2A, the cell numbers were lower than the group of Hek293 cells transfected with the FADD gene alone and higher than the group of Hek293 cells transfected with the FADD gene alone. The results correlated with the animal model as well. MT2A or pFADD gene knockdown in the animal model cancer size was minimized. Based on these data, we suggested that pFADD with MT2A might inhibit colorectal cancer apoptosis and increase cell proliferation.
Interesting observation was made with MT-II interacts with the p65 subunits of NF-κB, and MT-II has been found to be a positive regulator of NF-κB activity in this study. But some reports claim it has negative regulator of NF-κB (44), hereby in this study concludes and supporting an antioncogenic effect of MT-II in relation to NF-κB. The new thing in this study is resides in the identification of the pFADD with MT2A involved in NF-κB pathway as a critical mediator of increase cell proliferation leads to cancer formation, migration and invasion of Hek293 cells. By monitoring with gene knockdown it revealed that it’s a positive regulator of NF-κB pathway.
Finally taking together with our findings, the pFADD gene with MT2A can inhibit the apoptosis and influence proliferation, of colorectal cancer cells, and gene knock down by antisense sequence of MT2A and pFADD approaches which might swell the combination of deregulated proliferation and suppressed apoptosis. Because deregulated proliferation and inhibition of apoptosis lie at the heart of all tumor development, the information concerning the effect of pFADD and MT2A on Colo 205 cancer cell proliferation and apoptosis may provide new opportunities for target selection in designing new anti-cancer agents.
Acknowledgements
The authors thank Zheng Yun Zi and Zheng Wei at Nanjing University for their assistance with amending the manuscript.
Funding: Special thank goes to our funding agent, Chinese National Nature Science Foundation (Grant No. 30330530, 30425009).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All experimental protocols were approved under animal protocol number SYXK(Su)2009-0017 by the Animal Care and Use Committee of College of Life Sciences, Nanjing University, and animal welfare and treatment were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals (the Ministry of Science and Technology of China, 2006).
References
- Emmert-Buck MR, Gillespie JW, Paweletz CP, et al. An approach to proteomic analysis of human tumors. Mol Carcinog 2000;27:158-65. [Crossref] [PubMed]
- Bay BH, Jin R, Huang J, et al. Metallothionein as a prognostic biomarker in breast cancer. Exp Biol Med (Maywood) 2006;231:1516-21. [PubMed]
- Ferlay J, Shin HR, Bray F, et al. GLOBOCAN 2008 v2.0, Cancer incidence and mortality worldwide: IARC CancerBase No. 10. Available online: http://gco.iarc.fr/
- Watson AJ, Collins PD. Colon cancer: a civilization disorder. Dig Dis 2011;29:222-8. [Crossref] [PubMed]
- Toby GG, Golemis EA. Using the yeast interaction trap and other two-hybrid-based approaches to study protein-protein interactions. Methods 2001;24:201-17. [Crossref] [PubMed]
- Su T, Liu H, Lu S. Cloning and identification of cDNA fragments related to human esophageal cancer. Zhonghua Zhong Liu Za Zhi 1998;20:254-7. [PubMed]
- Marikar FM, Ma D, Ye J, et al. Expression of recombinant human FADD, preparation of its polyclonal antiserum and the application in immunoassays. Cell Mol Immunol 2008;5:471-4. [Crossref] [PubMed]
- Inoue N, Matsuda-Minehata F, Goto Y, et al. Molecular characteristics of porcine Fas-associated death domain (FADD) and procaspase-8. J Reprod Dev 2007;53:427-36. [Crossref] [PubMed]
- Hua ZC, Sohn SJ, Kang C, et al. A function of Fas-associated death domain protein in cell cycle progression localized to a single amino acid at its C-terminal region. Immunity 2003;18:513-21. [Crossref] [PubMed]
- Sato M, Kondoh M. Recent studies on metallothionein: protection against toxicity of heavy metals and oxygen free radicals. Tohoku J Exp Med 2002;196:9-22. [Crossref] [PubMed]
- Cherian MG, Jayasurya A, Bay BH. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res 2003;533:201-9. [Crossref] [PubMed]
- Thirumoorthy N, Manisenthil Kumar KT, Shyam Sundar A, et al. Metallothionein: an overview. World J Gastroenterol 2007;13:993-6. [Crossref] [PubMed]
- Klaassen CD, Liu J, Choudhuri S. Metallothionein: an intracellular protein to protect against cadmium toxicity. Annu Rev Pharmacol Toxicol 1999;39:267-94. [Crossref] [PubMed]
- Miles AT, Hawksworth GM, Beattie JH, et al. Induction, regulation, degradation, and biological significance of mammalian metallothioneins. Crit Rev Biochem Mol Biol 2000;35:35-70. [Crossref] [PubMed]
- Nielsen AE, Bohr A, Penkowa M. The Balance between Life and Death of Cells: Roles of Metallothioneins. Biomark Insights 2007;1:99-111. [PubMed]
- Coyle P, Philcox JC, Carey LC, et al. Metallothionein: the multipurpose protein. Cell Mol Life Sci 2002;59:627-47. [Crossref] [PubMed]
- Penkowa M. Metallothioneins are multipurpose neuroprotectants during brain pathology. FEBS J 2006;273:1857-70. [Crossref] [PubMed]
- Haq F, Mahoney M, Koropatnick J. Signaling events for metallothionein induction. Mutat Res 2003;533:211-26. [Crossref] [PubMed]
- Formigari A, Irato P, Santon A. Zinc, antioxidant systems and metallothionein in metal mediated-apoptosis: biochemical and cytochemical aspects. Comp Biochem Physiol C Toxicol Pharmacol 2007;146:443-59. [Crossref] [PubMed]
- Cherian MG, Kang YJ. Metallothionein and liver cell regeneration. Exp Biol Med (Maywood) 2006;231:138-44. [PubMed]
- Theocharis SE, Margeli AP, Klijanienko JT, et al. Metallothionein expression in human neoplasia. Histopathology 2004;45:103-18. [Crossref] [PubMed]
- Simpkins CO. Metallothionein in human disease. Cell Mol Biol (Noisy-le-grand) 2000;46:465-88. [PubMed]
- Jin R, Bay BH, Chow VT, et al. Significance of metallothionein expression in breast myoepithelial cells. Cell Tissue Res 2001;303:221-6. [Crossref] [PubMed]
- Cui Y, Wang J, Zhang X, et al. ECRG2, a novel candidate of tumor suppressor gene in the esophageal carcinoma, interacts directly with metallothionein 2A and links to apoptosis. Biochem Biophys Res Commun 2003;302:904-15. [Crossref] [PubMed]
- Zhao N, Wang J, Cui Y, et al. Induction of G1 cell cycle arrest and P15INK4b expression by ECRG1 through interaction with Miz-1. J Cell Biochem 2004;92:65-76. [Crossref] [PubMed]
- Kasperczyk H, La Ferla-Brühl K, Westhoff MA, et al. Betulinic acid as new activator of NF-kappaB: molecular mechanisms and implications for cancer therapy. Oncogene 2005;24:6945-56. [Crossref] [PubMed]
- Wang S, Li KJ, Lin XW, et al. Using c-Fos/c-Jun as hetero-dimer interaction model to optimize donor to acceptor concentration ratio range for three-filter fluorescence resonance energy transfer (FRET) measurement. J Micros 2012;248:58-65. [Crossref] [PubMed]
- Tchoghandjian A, Jennewein C, Eckhardt I, et al. Identification of non-canonical NF-κB signaling as a critical mediator of Smac mimetic-stimulated migration and invasion of glioblastoma cells. Cell Death Dis 2013;4:e564. [Crossref] [PubMed]
- Gordon GW, Berry G, Liang XH, et al. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys J 1998;74:2702-13. [Crossref] [PubMed]
- Youvan DC, Silva CM, Bylina EJ, et al. Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel cHeLating beads. Biotechnology et alia 1997;3:1-18.
- Sorkina T, Doolen S, Galperin E, et al. Oligomerization of dopamine transporters visualized in living cells by fluorescence resonance energy transfer microscopy. J Biol Chem 2003;278:28274-83. [Crossref] [PubMed]
- Erickson MG, Alseikhan BA, Peterson BZ, et al. Preassociation of calmodulin with voltage-gated Ca(2+) channels revealed by FRET in single living cells. Neuron 2001;31:973-85. [Crossref] [PubMed]
- Yap X, Tan HY, Huang J, et al. Over-expression of metallothionein predicts chemoresistance in breast cancer. J Pathol 2009;217:563-70. [Crossref] [PubMed]
- Carina D, Richard G. The Human Genome. New York: Nature/Palgrave Press, 2001.
- Jasny BR, Kennedy D. The human genome. Science 2001;291:1153. [Crossref] [PubMed]
- Eisenberg D, Marcotte EM, Xenarios I, et al. Protein function in the post-genomic era. Nature 2000;405:823-6. [Crossref] [PubMed]
- Chambers G, Lawrie L, Cash P, et al. Proteomics: a new approach to the study of disease. J Pathol 2000;192:280-8. [Crossref] [PubMed]
- Pandey A, Mann M. Proteomics to study genes and genomes. Nature 2000;405:837-46. [Crossref] [PubMed]
- McCluggage WG, Strand K, Abdulkadir A. Immunohistochemical localization of metallothionein in benign and malignant epithelial ovarian tumors. Int J Gynecol Cancer 2002;12:62-5. [Crossref] [PubMed]
- Ito T, Tashiro K, Muta S, et al. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A 2000;97:1143-7. [Crossref] [PubMed]
- Janssen AM, van Duijn W, Oostendorp-Van De Ruit MM, et al. Metallothionein in human gastrointestinal cancer. J Pathol 2000;192:293-300. [Crossref] [PubMed]
- Bouzourene H, Chaubert P, Gebhard S, et al. Role of metallothioneins in irradiated human rectal carcinoma. Cancer 2002;95:1003-8. [Crossref] [PubMed]
- Meijer C, Timmer A, De Vries EG, et al. Role of metallothionein in cisplatin sensitivity of germ-cell tumours. Int J Cancer 2000;85:777-81. [Crossref] [PubMed]
- Sakurai A, Hara S, Okano N, et al. Regulatory role of metallothionein in NF-kappaB activation. FEBS Lett 1999;455:55-8. [Crossref] [PubMed]


